Genealogy * Hermeneutics * Genetics * Philosophy * Anthropology * Shamanism * History * Esoterics * Myth * Mysticism
DRAGON GENES
Bloodline of Eden:
The Underground Stream
We began with a Key Concept of Psychosocial Genomics:
“Nothing, it seems turns on gene expression and brain plasticity as much as the presence of others of the same species!” ~Ernest Rossi, M.D.
Genetic Genealogy * Ethnogenetics * Electrogenetics *
Psychosocial Genomics
Psychosocial Genomics and Mind-Body Healing:
Recent research on neuroplasticity and psychosocial genomics lends compelling support to this perspective by elucidating mechanisms through which psychosocial forces shape neurobiology. Investigations of neuroplasticity demonstrate that the adult brain can continue to form novel neural connections and grow new neurons in response to learning or training even into old age. These findings are complemented by the contributions of psychosocial genomics, a field of scientific inquiry that explores the modulating effects of experience on gene expression. Findings from these new sciences provide external validation for the biopsychosocial perspective and offer important insights into the manifold means by which socioenvironmental experiences influence neurobiological structure and function across the life course.
What has psychosocial genomics have to do with mind-body healing? The answer is, "A lot." First, we need a few definitions so we can discuss this...
Definitions Psychosocial genomics can be defined as how personal consciousness, activities, and social interactions affect gene expression, and how gene expression affects these.
Another definition would be how psychological and social interactions and activities affect the genome in terms of gene expression and gene expression affects our psychology and social behaviors.
Rossi in his 2002 book, The Psychobiology of Gene Expression, defines psychosocial genomics as "how the subjective experiences of human consciousness, our perception of free will, and social dynamics can modulate gene expression, and visa versa."
Genome In the strict meaning to the word, the genome constitutes the entire set of an individual's genes. In a more dynamic sense, it encompasses not only all the genes, but also the way those genes interact with each other.
Gene Expression It is not just a matter of what genes an individual has, but what genes are being expressed, when they are being expressed, and to what degree or how much they are being expressed. "Express" here refers to the protein or RNA that is encoded by a gene. For example, for protein coding genes, how much of that protein is actually being produced? That is gene expression is regulated.
A gene is not just simply sitting there pumping out proteins (or RNA) but is dynamically adjusting its output based on feedback from the internal and external environments.
Dynamic Dance of Gene Expression From recent findings in the neurosciences and functional genomics we are beginning to understand how the interplay between behavioral state-related gene expression (a.k.a. nature) and activity-dependent gene expression (a.k.a. nurture) bring about healing through neurogenesis and learning.
Functional genomics has to do with how the genes in the genome function in response to the environment. During embryonic development for example, we see cascades of genes being turned on and off to varying degrees. Even from such simple interactions of touch, we see whole communities of genes responding. So when someone comes up and gives you a hug, rubs your back, etc., your genes respond to that. It is specific genes that respond however.
Mind-body healing is about neurogenesis or brain growth. For neurogenesis to occur, the genes involved have to produce their proteins and have them available for nerve cells (neurons) to use.
As reviewed in the neurogenesis webpage, three things stimulate brain growth: novelty, a variable environment, and exercise. One of the most powerful of these for psychosocial genomics is what Rossi (2002) refers to as the novelty-numinosum-neurogenesis process.
Novelty-Numinosum-Neurogenesis Novelty (that is doing or experiencing something really different) can lead to numinous experiences and numinous experiences are novel.
Numinous refers to supernatural or mystical experience. They are filled with the presence of the sacred or divine. Maslow's Peak Experiences are of this nature. Gerald May (see Will and Spirit: A Contemplative Psychology) refers to these as unitive experiences. They have also been called religious conversion experiences. Buddhism would refer to them as enlightenment experiences (satori or samadi). To a lesser degree they are related to the Aha-experiences that occur during psychotherapy.
These experience are novel and powerful and can generate neurogenesis and healing. They can result in major reorganization of our mind's inner world. This can be life changing.
An Introduction to Psychosocial Genomics: How the body speaks to us about the effects of non-invasive processes such as therapeutic hypnosis
Hill, R. 2010 . Australian Journal of Clinical Hypnotherapy and Hypnosis 31 (1), pp. 5-16
Until recently, therapeutic hypnosis and the mind-body therapies have had little capacity to make empirical measurements of their benefit with an objective, common standard of reference. Among many therapies such as the talk therapy (e.g. cognitive-behaviour psychotherapy, counselling, etc), energy therapies(e.g. reiki, acupuncture,homeopathy) and massage therapies (e.g. remedial, Shiatsu, trigger point), for example, there Is no general agreed upon measure by which they can all be evaluated. Mostly, studies have depended upon phenomenological reporting of improvement In sense of well-being, affect, and other perceptions of health improvement. By its nature, however, phenomenological reporting has a high level of subjectivity. Although subjectivity In itself is not a reason to discount such evidence, it would certainly be beneficial if there was a way of objectively measuring responses In the body. After several decades of prescient study, Ernest Rossi (1986, 2002, 2004, 2007) has found a common, objective measurement, which provides a yardstick that transcends personal subjectivity. He proposes that the new DNA microarray technology provides objective evidence for evaluating all the mindbody therapies. This new field of empirical investigation, which he calls, “Psychosocial Genomics,” measures changes in the deep psychoblological process of “activity or experience-dependent gene expression and brain plasticity”associated with creativity and psychotherapy. By examining which genes are turned on or off before and after therapeutic hypnosis, Rossi has been able to establish that genes beneficial to health and well being are expressed in the hours and days following the use of his Activity-Dependent Mirror Hand Protocol for therapeutic hypnosis (Rossi, lannotti et al., 2008). This paper outlines the conceptions and research of Ernest Rossi into the deep psychoblological basis of consciousness and the nascent possibilities for future research in therapeutic hypnosis and all the mind-body therapies.
1. Rossi, E. (2002). The Psychobiology of Gene Expression: Neuroscience and Neurogenesis in Hypnosis and the Healing arts. NY: W. W. Norton.
2. Rossi, E. (2004). A Discourse with Our Genes: The psychosocial and cultural genomics of therapeutic hypnosis and psychotherapy. Available in English and Italian. (ISBN –89396-01-6) San Lorenzo Maggiore, Italy: Editris s.a.s. Phoenix, Arizona: Zeig, Tucker & Theisen.
3. Rossi, E. (2004). Art, Beauty and Truth: The Psychosocial Genomics of Consciousness, Dreams, and Brain Growth in Psychotherapy and Mind-Body Healing. Annals of the American Psychotherapy Association, 7, 10-17.
4. Rossi, E. (2004). Sacred Spaces and Places in Healing Dreams: Gene expression and brain growth in rehabilitation: Psychological Perspectives, 47, 48-63.
5. Rossi, E. (2005). Creativity and the Nature of the Numinosum: The Psychosocial Genomics of Jung’s Transcendent Function in Art, Science, Spirit, & Psychotherapy. Spring, 72, 313-337.1.
6. Rossi, E. (2007). The Breakout Heuristic: The New Neuroscience of Mirror Neurons, Consciousness and Creativity in Human Relationships: Selected Papers of Ernest Lawrence Rossi. Phoenix, Arizona: The Milton H. Erickson Foundation Press.
7. Rossi, E., Iannotti, S., Cozzolino, M., Castiglione, S., Cicatelli, A. & Rossi, K. (2008). A pilot study of positive expectations and focused attention via a new protocol for therapeutic hypnosis assessed with DNA microarrays: The creative psychosocial genomic healing experience. Sleep and Hypnosis: An International Journal of Sleep, Dream, and Hypnosis, 10:2, 39-44.
The Psychosocial Genomics of Therapeutic Hypnosis, Psychotherapy, and Rehabilitation
Ernest Lawrence Rossi
http://www.asch.net/portals/0/journallibrary/articles/ajch-51/51-3/rossi51-3.pdf
The Bioinformatics of Psychosocial Genomics in Alternative and Complementary Medicine
E. Rossi
http://www.ernestrossi.com/ernestrossi/keypapers/PG%20Bioinformatics%20Psychosocial%20Genomics%202003.pdf
Gene Expression, Neurogenesis, and Healing: Psychosocial Genomics of Therapeutic Hypnosis
Ernest L. Rossi, Los Osos, California
http://ernestrossi.com/ernestrossi/keypapers/PG%20Gene%20Expression%20Nuerogenesis%20and%20Healing%202003.PDF
Neüroplasticity, Psychosocial Genomics, and the Biopsychosocial Paradigm in the 21st Century.
Eric L. Garland and Matthew Owen Howard
http://integralsocialwork.com/documents/neuroplasticity.pdf
Recent research on neuroplasticity and psychosocial genomics lends compelling support to this perspective by elucidating mechanisms through which psychosocial forces shape neurobiology. Investigations of neuroplasticity demonstrate that the adult brain can continue to form novel neural connections and grow new neurons in response to learning or training even into old age. These findings are complemented by the contributions of psychosocial genomics, a field of scientific inquiry that explores the modulating effects of experience on gene expression. Findings from these new sciences provide external validation for the biopsychosocial perspective and offer important insights into the manifold means by which socioenvironmental experiences influence neurobiological structure and function across the life course.
What has psychosocial genomics have to do with mind-body healing? The answer is, "A lot." First, we need a few definitions so we can discuss this...
Definitions Psychosocial genomics can be defined as how personal consciousness, activities, and social interactions affect gene expression, and how gene expression affects these.
Another definition would be how psychological and social interactions and activities affect the genome in terms of gene expression and gene expression affects our psychology and social behaviors.
Rossi in his 2002 book, The Psychobiology of Gene Expression, defines psychosocial genomics as "how the subjective experiences of human consciousness, our perception of free will, and social dynamics can modulate gene expression, and visa versa."
Genome In the strict meaning to the word, the genome constitutes the entire set of an individual's genes. In a more dynamic sense, it encompasses not only all the genes, but also the way those genes interact with each other.
Gene Expression It is not just a matter of what genes an individual has, but what genes are being expressed, when they are being expressed, and to what degree or how much they are being expressed. "Express" here refers to the protein or RNA that is encoded by a gene. For example, for protein coding genes, how much of that protein is actually being produced? That is gene expression is regulated.
A gene is not just simply sitting there pumping out proteins (or RNA) but is dynamically adjusting its output based on feedback from the internal and external environments.
Dynamic Dance of Gene Expression From recent findings in the neurosciences and functional genomics we are beginning to understand how the interplay between behavioral state-related gene expression (a.k.a. nature) and activity-dependent gene expression (a.k.a. nurture) bring about healing through neurogenesis and learning.
Functional genomics has to do with how the genes in the genome function in response to the environment. During embryonic development for example, we see cascades of genes being turned on and off to varying degrees. Even from such simple interactions of touch, we see whole communities of genes responding. So when someone comes up and gives you a hug, rubs your back, etc., your genes respond to that. It is specific genes that respond however.
Mind-body healing is about neurogenesis or brain growth. For neurogenesis to occur, the genes involved have to produce their proteins and have them available for nerve cells (neurons) to use.
As reviewed in the neurogenesis webpage, three things stimulate brain growth: novelty, a variable environment, and exercise. One of the most powerful of these for psychosocial genomics is what Rossi (2002) refers to as the novelty-numinosum-neurogenesis process.
Novelty-Numinosum-Neurogenesis Novelty (that is doing or experiencing something really different) can lead to numinous experiences and numinous experiences are novel.
Numinous refers to supernatural or mystical experience. They are filled with the presence of the sacred or divine. Maslow's Peak Experiences are of this nature. Gerald May (see Will and Spirit: A Contemplative Psychology) refers to these as unitive experiences. They have also been called religious conversion experiences. Buddhism would refer to them as enlightenment experiences (satori or samadi). To a lesser degree they are related to the Aha-experiences that occur during psychotherapy.
These experience are novel and powerful and can generate neurogenesis and healing. They can result in major reorganization of our mind's inner world. This can be life changing.
An Introduction to Psychosocial Genomics: How the body speaks to us about the effects of non-invasive processes such as therapeutic hypnosis
Hill, R. 2010 . Australian Journal of Clinical Hypnotherapy and Hypnosis 31 (1), pp. 5-16
Until recently, therapeutic hypnosis and the mind-body therapies have had little capacity to make empirical measurements of their benefit with an objective, common standard of reference. Among many therapies such as the talk therapy (e.g. cognitive-behaviour psychotherapy, counselling, etc), energy therapies(e.g. reiki, acupuncture,homeopathy) and massage therapies (e.g. remedial, Shiatsu, trigger point), for example, there Is no general agreed upon measure by which they can all be evaluated. Mostly, studies have depended upon phenomenological reporting of improvement In sense of well-being, affect, and other perceptions of health improvement. By its nature, however, phenomenological reporting has a high level of subjectivity. Although subjectivity In itself is not a reason to discount such evidence, it would certainly be beneficial if there was a way of objectively measuring responses In the body. After several decades of prescient study, Ernest Rossi (1986, 2002, 2004, 2007) has found a common, objective measurement, which provides a yardstick that transcends personal subjectivity. He proposes that the new DNA microarray technology provides objective evidence for evaluating all the mindbody therapies. This new field of empirical investigation, which he calls, “Psychosocial Genomics,” measures changes in the deep psychoblological process of “activity or experience-dependent gene expression and brain plasticity”associated with creativity and psychotherapy. By examining which genes are turned on or off before and after therapeutic hypnosis, Rossi has been able to establish that genes beneficial to health and well being are expressed in the hours and days following the use of his Activity-Dependent Mirror Hand Protocol for therapeutic hypnosis (Rossi, lannotti et al., 2008). This paper outlines the conceptions and research of Ernest Rossi into the deep psychoblological basis of consciousness and the nascent possibilities for future research in therapeutic hypnosis and all the mind-body therapies.
1. Rossi, E. (2002). The Psychobiology of Gene Expression: Neuroscience and Neurogenesis in Hypnosis and the Healing arts. NY: W. W. Norton.
2. Rossi, E. (2004). A Discourse with Our Genes: The psychosocial and cultural genomics of therapeutic hypnosis and psychotherapy. Available in English and Italian. (ISBN –89396-01-6) San Lorenzo Maggiore, Italy: Editris s.a.s. Phoenix, Arizona: Zeig, Tucker & Theisen.
3. Rossi, E. (2004). Art, Beauty and Truth: The Psychosocial Genomics of Consciousness, Dreams, and Brain Growth in Psychotherapy and Mind-Body Healing. Annals of the American Psychotherapy Association, 7, 10-17.
4. Rossi, E. (2004). Sacred Spaces and Places in Healing Dreams: Gene expression and brain growth in rehabilitation: Psychological Perspectives, 47, 48-63.
5. Rossi, E. (2005). Creativity and the Nature of the Numinosum: The Psychosocial Genomics of Jung’s Transcendent Function in Art, Science, Spirit, & Psychotherapy. Spring, 72, 313-337.1.
6. Rossi, E. (2007). The Breakout Heuristic: The New Neuroscience of Mirror Neurons, Consciousness and Creativity in Human Relationships: Selected Papers of Ernest Lawrence Rossi. Phoenix, Arizona: The Milton H. Erickson Foundation Press.
7. Rossi, E., Iannotti, S., Cozzolino, M., Castiglione, S., Cicatelli, A. & Rossi, K. (2008). A pilot study of positive expectations and focused attention via a new protocol for therapeutic hypnosis assessed with DNA microarrays: The creative psychosocial genomic healing experience. Sleep and Hypnosis: An International Journal of Sleep, Dream, and Hypnosis, 10:2, 39-44.
The Psychosocial Genomics of Therapeutic Hypnosis, Psychotherapy, and Rehabilitation
Ernest Lawrence Rossi
http://www.asch.net/portals/0/journallibrary/articles/ajch-51/51-3/rossi51-3.pdf
The Bioinformatics of Psychosocial Genomics in Alternative and Complementary Medicine
E. Rossi
http://www.ernestrossi.com/ernestrossi/keypapers/PG%20Bioinformatics%20Psychosocial%20Genomics%202003.pdf
Gene Expression, Neurogenesis, and Healing: Psychosocial Genomics of Therapeutic Hypnosis
Ernest L. Rossi, Los Osos, California
http://ernestrossi.com/ernestrossi/keypapers/PG%20Gene%20Expression%20Nuerogenesis%20and%20Healing%202003.PDF
Neüroplasticity, Psychosocial Genomics, and the Biopsychosocial Paradigm in the 21st Century.
Eric L. Garland and Matthew Owen Howard
http://integralsocialwork.com/documents/neuroplasticity.pdf
Awakening
Bright sun
Bursting forth
Into the dawn
Of a brand new day
Golden rays
Streak across the sky
Blanketing the land
With newfound warmth
Manifold lifeforms
Arise from their slumber
To the infinite possibilities
Of this grand awakening
Infinite choice
Presented to all
With the dawn
Of this new day
Do we choose
To let wounds of the past
And fears of the future
Determine our way?
Or do we choose
To stir and awaken
The limitless glories
Within and around us?
May we transcend
Past and future shadows
To dance with the golden rays
Of the newborn sun
--Fred Burks
Bright sun
Bursting forth
Into the dawn
Of a brand new day
Golden rays
Streak across the sky
Blanketing the land
With newfound warmth
Manifold lifeforms
Arise from their slumber
To the infinite possibilities
Of this grand awakening
Infinite choice
Presented to all
With the dawn
Of this new day
Do we choose
To let wounds of the past
And fears of the future
Determine our way?
Or do we choose
To stir and awaken
The limitless glories
Within and around us?
May we transcend
Past and future shadows
To dance with the golden rays
Of the newborn sun
--Fred Burks
Biophotons Synchronize the Bodymind
EVERY living cell transmits and receives energy across the electromagnetic spectrum from radio waves through infrared, visible and ultraviolet wavelengths. Biophotons provide the means for near instantaneous communication throughout our bodies, and create an intricate web of energy within ALL living systems. It is now well established that all living systems emit a weak but permanent photon flux in the visible and ultraviolet range. This biophoton emission is correlated with many, if not all, biological and physiological functions. There are indications of a hitherto-overlooked information channel within the living system. Biophotons may trigger chemical reactivity in cells, growth control, differentiation and intercellular communication, i.e. biological rhythms.
Biophotons are weak emissions of light radiated from the cells of all living things. A photon is a single particle of light. Plants, animals and humans generate up to 100 photons per second, per .15 square inches (1 sq. centimeter) of surface area. The light is too faint to be seen by the naked eye, but biophotons have been detected and verified using photomultiplier tubes.
According to a leading researcher of biophotons, German biophysicist Fritz-Albert Popp, light is constantly being absorbed and remitted by DNA molecules within each cell's nucleus. These biophotons create a dynamic, coherent web of light. A system that could be responsible for chemical reactions within the cells, cellular communication throughout the organism, and the overall regulation of the biological system, including embryonic development into a predetermined form.
The laser-like coherence of the biophoton field is a significant attribute, making it a prime candidate for exchanging information in a highly functional, efficient and cooperative fashion, lending credence to the idea that it may be the intelligence factor behind the biological processes. An aspect of, or cousin to consciousness, though this remains speculative.
This light emission is an expression of the functional state of the living organism and it's measurement, therefore can be used to assess this state. Cancer cells and healthy cells of the same type, for instance, can be discriminated by typical differences in Biophoton emission. After an initial decade and a half of basic research on this discovery, biophysicists of various European and Asian countries are now exploring the MANY interesting applications, which range across such diverse fields as cancer research, non-invasive early medical diagnosis, food and water quality testing, chemical and electromagnetic contamination testing, cell communication, and various applications in biotechnology.
According to the Biophoton theory developed on the base of these discoveries the Biophoton light is stored in the cells of the organism - more precisely, in the DNA molecules of their nuclei - and a dynamic web of light constantly released and absorbed by the DNA may connect cell organelles, cells, tissues, and organs within the body and serve as the organism's main communication network and as the principal regulating instance for ALL life processes. The processes of morphogenesis, growth, differentiation and regeneration are also explained by the structuring and regulating activity of the coherent Biophoton field. The holographic Biophoton field of the brain and the nervous system, and maybe even that of the WHOLE organism, may also be basis of memory and other phenomena of consciousness, as postulated by neurophysiologist Karl Pribram an others. The consciousness-like coherence properties of the Biophoton field are closely related to it's base in the properties of the physical vacuum and indicate it's possible role as an interface to the non-physical realms of mind, psyche and consciousness.
The discovery of Biophoton emission also lends scientific support to some unconventional methods of healing based on concepts of homeostasis (self-regulation of the organism), such as various somatic therapies, homeopathy and acupuncture. The "chi" energy flowing in our bodies' energy channels (meridians), which according to Traditional Chinese Medicine regulates our body functions may be related to node lines of the organism's Biophoton field. The "prana" of Indian Yoga physiology may be a similar regulating energy force, that has a basis in WEAK, coherent electromagnetic Biophoton fields.
EVERY living cell transmits and receives energy across the electromagnetic spectrum from radio waves through infrared, visible and ultraviolet wavelengths. Biophotons provide the means for near instantaneous communication throughout our bodies, and create an intricate web of energy within ALL living systems. It is now well established that all living systems emit a weak but permanent photon flux in the visible and ultraviolet range. This biophoton emission is correlated with many, if not all, biological and physiological functions. There are indications of a hitherto-overlooked information channel within the living system. Biophotons may trigger chemical reactivity in cells, growth control, differentiation and intercellular communication, i.e. biological rhythms.
Biophotons are weak emissions of light radiated from the cells of all living things. A photon is a single particle of light. Plants, animals and humans generate up to 100 photons per second, per .15 square inches (1 sq. centimeter) of surface area. The light is too faint to be seen by the naked eye, but biophotons have been detected and verified using photomultiplier tubes.
According to a leading researcher of biophotons, German biophysicist Fritz-Albert Popp, light is constantly being absorbed and remitted by DNA molecules within each cell's nucleus. These biophotons create a dynamic, coherent web of light. A system that could be responsible for chemical reactions within the cells, cellular communication throughout the organism, and the overall regulation of the biological system, including embryonic development into a predetermined form.
The laser-like coherence of the biophoton field is a significant attribute, making it a prime candidate for exchanging information in a highly functional, efficient and cooperative fashion, lending credence to the idea that it may be the intelligence factor behind the biological processes. An aspect of, or cousin to consciousness, though this remains speculative.
This light emission is an expression of the functional state of the living organism and it's measurement, therefore can be used to assess this state. Cancer cells and healthy cells of the same type, for instance, can be discriminated by typical differences in Biophoton emission. After an initial decade and a half of basic research on this discovery, biophysicists of various European and Asian countries are now exploring the MANY interesting applications, which range across such diverse fields as cancer research, non-invasive early medical diagnosis, food and water quality testing, chemical and electromagnetic contamination testing, cell communication, and various applications in biotechnology.
According to the Biophoton theory developed on the base of these discoveries the Biophoton light is stored in the cells of the organism - more precisely, in the DNA molecules of their nuclei - and a dynamic web of light constantly released and absorbed by the DNA may connect cell organelles, cells, tissues, and organs within the body and serve as the organism's main communication network and as the principal regulating instance for ALL life processes. The processes of morphogenesis, growth, differentiation and regeneration are also explained by the structuring and regulating activity of the coherent Biophoton field. The holographic Biophoton field of the brain and the nervous system, and maybe even that of the WHOLE organism, may also be basis of memory and other phenomena of consciousness, as postulated by neurophysiologist Karl Pribram an others. The consciousness-like coherence properties of the Biophoton field are closely related to it's base in the properties of the physical vacuum and indicate it's possible role as an interface to the non-physical realms of mind, psyche and consciousness.
The discovery of Biophoton emission also lends scientific support to some unconventional methods of healing based on concepts of homeostasis (self-regulation of the organism), such as various somatic therapies, homeopathy and acupuncture. The "chi" energy flowing in our bodies' energy channels (meridians), which according to Traditional Chinese Medicine regulates our body functions may be related to node lines of the organism's Biophoton field. The "prana" of Indian Yoga physiology may be a similar regulating energy force, that has a basis in WEAK, coherent electromagnetic Biophoton fields.
The Biophoton © 2002 Elizabeth Bauer
The term "bio" in biophotons was introduced to point out the classification of photons being emitted from a biological source. This phenomenon was characterized by measuring single photons. This indicated that the biophoton is subject to quantum optics rather than classical physics. Biophotons are photons emitted spontaneously by all living systems. Biophotons are characterized by delayed luminescence and are associated with biological systems hence the name biophotons as distinct from photons which are normally associated with inanimate physical systems.
The biophoton phenomenon is not confined to "thermal" radiation in the infrared range. It is well known that biophotons are emitted also in the range from visible up to the UV ranges of the electromagnetic spectrum. The intensity of biophotons can be registered from a few photons per second in a square centimeter surface area to several hundred photons per second in a square centimeter from every living system.
"The high degree of coherence of biophotons elucidates the universal phenomenon of biological systems — coherence of biophotons is responsible for the information transfer within and between cells. This answers the crucial question of intra and extracellular biocommunication, including the regulation of metabolic activities of cells, growth, differentiation and evolutionary development." — F.A. Popp, 1999
Biological systems are governed by the interactions of energy fields that are electromagnetic by nature. These energy fields are emitted by the biophotons derived from the biological matter. The energy fields dirigate the location and activity of matter, while matter provides the boundary for the energy fields. Thus, we define the correlations between energy and matter.
"An ordinary cell has a diameter of approximately 10 -3 cm. Inside the cell there is a rather high metabolic activity of about 10 5 reactions per second. For every reaction the suitable activation energy (in the range from microwaves to the ultraviolet) is necessary to establish the formation of the transitional state complex that finally decays into chemical products. Biochemical reactions take place in a way that a photon is borrowed from the surrounding electromagnetic bath, then, it excites the transition state complex and finally returns to the equilibrium states of the surroundings, becoming available for the next reaction. The single photon may suffice to trigger about 10 9 reactions per second. The reaction is directed in a way that it delivers the right activation energy as well as the right momentum at the right time to the right place. Thus, a surprisingly low photon intensity may suffice to trigger all of the chemical reactions in a cell. Despite the low intensities, at any given instant at least 10 10 to 10 40 more photons are available than under thermal equilibrium conditions.
A temperature increase of 10° doubles the photon density of a thermal field under physiological conditions resulting in a doubling of the reaction rate."
— F.A. Popp, 1999
Biophotons also have a characteristic frequency that defines their resonance patterns and energy distribution. The study of these frequencies and resonance patterns is vital in the understanding the omnifarious electromagnetic spectrum and how its energies can be harnessed and effectively utilized therapeutically, in biological systems. http://www.biophotonicsresearchinstitute.com/ShortArticles.htm
The term "bio" in biophotons was introduced to point out the classification of photons being emitted from a biological source. This phenomenon was characterized by measuring single photons. This indicated that the biophoton is subject to quantum optics rather than classical physics. Biophotons are photons emitted spontaneously by all living systems. Biophotons are characterized by delayed luminescence and are associated with biological systems hence the name biophotons as distinct from photons which are normally associated with inanimate physical systems.
The biophoton phenomenon is not confined to "thermal" radiation in the infrared range. It is well known that biophotons are emitted also in the range from visible up to the UV ranges of the electromagnetic spectrum. The intensity of biophotons can be registered from a few photons per second in a square centimeter surface area to several hundred photons per second in a square centimeter from every living system.
"The high degree of coherence of biophotons elucidates the universal phenomenon of biological systems — coherence of biophotons is responsible for the information transfer within and between cells. This answers the crucial question of intra and extracellular biocommunication, including the regulation of metabolic activities of cells, growth, differentiation and evolutionary development." — F.A. Popp, 1999
Biological systems are governed by the interactions of energy fields that are electromagnetic by nature. These energy fields are emitted by the biophotons derived from the biological matter. The energy fields dirigate the location and activity of matter, while matter provides the boundary for the energy fields. Thus, we define the correlations between energy and matter.
"An ordinary cell has a diameter of approximately 10 -3 cm. Inside the cell there is a rather high metabolic activity of about 10 5 reactions per second. For every reaction the suitable activation energy (in the range from microwaves to the ultraviolet) is necessary to establish the formation of the transitional state complex that finally decays into chemical products. Biochemical reactions take place in a way that a photon is borrowed from the surrounding electromagnetic bath, then, it excites the transition state complex and finally returns to the equilibrium states of the surroundings, becoming available for the next reaction. The single photon may suffice to trigger about 10 9 reactions per second. The reaction is directed in a way that it delivers the right activation energy as well as the right momentum at the right time to the right place. Thus, a surprisingly low photon intensity may suffice to trigger all of the chemical reactions in a cell. Despite the low intensities, at any given instant at least 10 10 to 10 40 more photons are available than under thermal equilibrium conditions.
A temperature increase of 10° doubles the photon density of a thermal field under physiological conditions resulting in a doubling of the reaction rate."
— F.A. Popp, 1999
Biophotons also have a characteristic frequency that defines their resonance patterns and energy distribution. The study of these frequencies and resonance patterns is vital in the understanding the omnifarious electromagnetic spectrum and how its energies can be harnessed and effectively utilized therapeutically, in biological systems. http://www.biophotonicsresearchinstitute.com/ShortArticles.htm
There is a traditional DNA based genetic code. Nevertheless, I have found the dynamic corollaries of the genetic code. This second code governs energy exchange and is the electronic receiver and transmitter of DNA. When it gets out of alignment, it can no longer exchange energy and we die. Dead things have only the first code; live things have both codes.
When seeds get wet and warm they induce the onset of the second code. The first code stores traits (protein synthesis). The second code expresses or suppresses traits (protein synthesis). The second code is a musical program of repeated phrases which re-phrases combinations throughout life. It requires alignment of the nucleosomes for transmission. These are specially coiled chromatin. They form a series inductance electronic device and send 50 milivolt signals to the catalytic centers of certain enzymes at ultra-low resonant frequencies. They depend on nucleosome inductance and alignment for charge efflux. Nucleosome alignment becomes critical and is detuned by competing paramagnetic centers at higher frequencies. Our compounds re-introduce the signal devices to mimic the stage in the electronic musical phrase.
When someone says there's a change in the DNA, please be aware that the context implies a change in the genomic sequence. This is always toxic. Much of conventional chemotherapy attempts to alter and break DNA. The things that do are mutagenic and not therapeutic. We do not do that. We do something different. By re-connecting the DNA electronically and thermodynamically, we tune the channel and are greeted by pulses of a musical nature! Transferring the charge in and out of DNA with the palladium lipoic complex changes the DNA charge and the charge on the cell membranes. This happens already in a normal cell in the same specific range. Induction of this normalization charge in tumors provides a novel therapeutic concept. http://newsgrist.typepad.com/electrogenetics/notes-toward-a-corollary-genetic-code.html
When seeds get wet and warm they induce the onset of the second code. The first code stores traits (protein synthesis). The second code expresses or suppresses traits (protein synthesis). The second code is a musical program of repeated phrases which re-phrases combinations throughout life. It requires alignment of the nucleosomes for transmission. These are specially coiled chromatin. They form a series inductance electronic device and send 50 milivolt signals to the catalytic centers of certain enzymes at ultra-low resonant frequencies. They depend on nucleosome inductance and alignment for charge efflux. Nucleosome alignment becomes critical and is detuned by competing paramagnetic centers at higher frequencies. Our compounds re-introduce the signal devices to mimic the stage in the electronic musical phrase.
When someone says there's a change in the DNA, please be aware that the context implies a change in the genomic sequence. This is always toxic. Much of conventional chemotherapy attempts to alter and break DNA. The things that do are mutagenic and not therapeutic. We do not do that. We do something different. By re-connecting the DNA electronically and thermodynamically, we tune the channel and are greeted by pulses of a musical nature! Transferring the charge in and out of DNA with the palladium lipoic complex changes the DNA charge and the charge on the cell membranes. This happens already in a normal cell in the same specific range. Induction of this normalization charge in tumors provides a novel therapeutic concept. http://newsgrist.typepad.com/electrogenetics/notes-toward-a-corollary-genetic-code.html
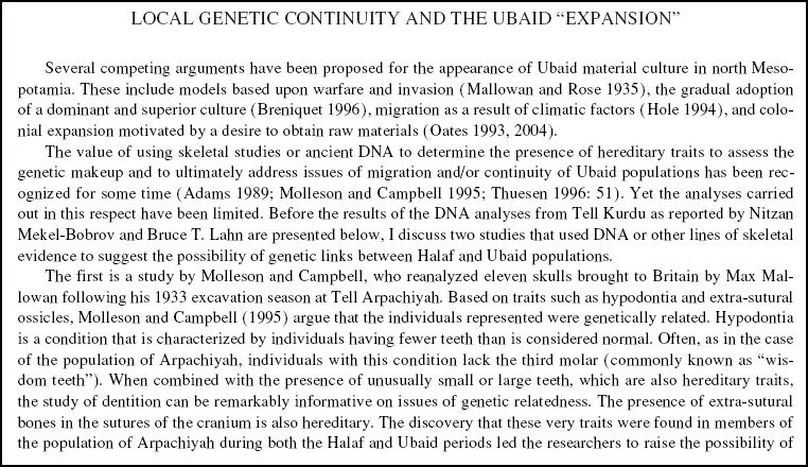
Ethnogenesis (from the Greek ethnos ἔθνος, "group of people" or "nation", and genesis γένεσις, "origin, birth", pl. ethnogeneses) is the process by which a group of human beings comes to be understood or to understand themselves as ethnically distinct from the wider social landscape from which their grouping emerges. Genetics is proving what legends could only allude to in legend and symbolic form...winged dragon genes.
Enki's philosophy manifests and explains itself in early Mesopotamian and Egyptian thought: the true creator of the universe was manifest within nature, and that nature enveloped both the Anunnaki, and the humans. Nature, as the Great Mother, was still supreme, despite any patriarchal scheme to the contrary. Admittedly, Enki’s claim of his birthright, the one being based on a matrilineal succession -- essentially the mitochondria DNA link, which is wholly passed through the female line -- was in Enki’s best interests. But Enki was also the maternal grandfather who came to the aid of Inanna when things went badly during her Descent into the Underworld. With the arrival of Enlil, however, who in his best interests must demean the matriarchal line of succession, and thus nature itself -- everything changed. The Great Mother was dethroned and replaced by a supreme male (as opposed to a male consort for the Queen). The idea of cooperation -- as exemplified by the council of Anunnaki making cooperative decisions -- was quickly replaced by competition, and harmony was forsaken in favor of subservience. The supreme god became abstract, and any physical connection with human or nature was lost -- and thus the link between nature and human also destroyed.
When Enlil hit town, there was a whole new deal put into effect. The new regime was based wholly on the utmost fear of Enlil, who instigated the great Flood without warning the humans, or making any attempt to save them. He facilitated the invasion and destruction of civilized Sumer. Here was a dictatorial deity who spared no mercy for those who did not comply with his authority.
When Enlil hit town, there was a whole new deal put into effect. The new regime was based wholly on the utmost fear of Enlil, who instigated the great Flood without warning the humans, or making any attempt to save them. He facilitated the invasion and destruction of civilized Sumer. Here was a dictatorial deity who spared no mercy for those who did not comply with his authority.
The Family IS Who You Are
Authenticity and legitimacy are based on factual history and the power of sacred history.
SayyiDNA means 'Our Master'. The na on the end attributes the word master to one's-self.
Plastic Gene-Expression: Ernest Rossi (1999; 2002) has developed a pertinent
creativity hypothesis: “Enriching life experiences that evoke psychobiological
arousal with positive fascination and focused attention
during creative
moments of art, music, dance, drama, humor, spirituality, numinosity, awe, joy,
expectation, and social rituals can evoke immediate early gene protein cascades
to optimize
brain growth, mindbody communication, and healing.”
brain growth, mindbody communication, and healing.”
Ancestral Discovery Method: http://en.wikipedia.org/wiki/Autosome
About Genetic Genealogy
Genetic genealogy is another tool for the genealogist's toolbox. It is the use of genetics to study genealogy, the relationship between individuals.
There are at least four types of genealogical DNA testing, including Y-chromosome, X-chromosome, mtDNA, and autosomal DNA test.
Y-chromosome:
The Y chromosome is only found in males, who have one Y chromosome and one X chromosome. The Y chromosome is composed of 58 million base pairs and contains 83 genes which code for only 23 proteins.
During meiosis (preparation of cells for reproductive purposes) the Y chromosome and the X chromosome only recombine at the very ends of the chromosomes (roughly 5%) in a region called the telomeres. Most of the Y chromosomes are located in this region, meaning that they are shared between both sex chromosomes.
The lack of recombination means that the Y chromosome is passed down from father to son without significant change. Over long periods of time the chromosome begins to accumulate mutations that are typically silent and have no impact on the carrier. These mutations, however, are useful for genealogical purposes “they can be used to analyze the relationships between populations and individuals.
When a Y chromosome is submitted for analysis, short segments of DNA known as short tandem repeats (STRs) are sequenced. STRs are also known by their DYS number (DNA Y-chromosome Segment number). The number of repeats can change over time and are passed on from father and son, giving scientists the ability to trace paternal lineages. STR analysis typically provides a person's haplotype, although they are often used to suggest which haplogroup an individual matches.
Some genetics tests also analyze SNPs (Single Nucleotide Polymorphisms), single nucleotide changes in the DNA sequence. SNPs are typically identified using enzymes that cleave DNA – a change in the DNA sequence can prevent the enzyme from cleaving the DNA, creating a pattern that can easily be analyzed. SNP analysis typically provides a person's haplogroup.
Most Y chromosome tests examine between 12 and 67 STR markers, but many more are known and could eventually be used for relationship analysis. Each STR has a numerical value. DYS392, for example, has a value of 11, 12, 13, 14, 15, of 16, with 13 being the most common. The numerical values for all the STRs tested can then be compared to the thousands of other numerical sets available in DNA databases. If two people have identical numerical values, it is more likely that they have a recent common ancestor. Statistical analysis can be used to calculate the number of generations to the most common recent ancestor (MRCA).
X-chromosome:
The X chromosome, found in both males and females, is more than 153 million base pairs and contains roughly 1000 genes. Females have two X chromosomes while males have just one.
The use of X chromosomes to study genealogical relationships is still relatively new. The X chromosome, just like the Y, contains STRs, called X-STRs. The problem with studying X-STRs is that the entire X chromosome undergoes recombination during meiosis. In other words, in females the two X chromosomes randomly swap information and genes. Family Tree DNA, one of the leaders of X-STR research (using methods developed by DNA-Fingerprint), uses “haplotype blocks, or regions of X-STRs that are inherited intact over several generations.
A male's X chromosome is inherited from his mother and is a mixture of her two X chromosomes, one from her mother and one from her father. It is therefore a mixture of the maternal grandparent's X chromosomes.
A female inherits one X chromosome from each of her two parents. The X chromosome from her father is passed on from his mother is a mixture of her parents (the paternal great-grandparent's) DNA, while the X chromosome from the female's mother is a mixture of her parent’s (the maternal grandparent's) DNA.
mtDNA:
Mitochondrial DNA (mtDNA) is a small circle of DNA that is located inside a small organelle found inside our cells, the mitochondria. mtDNA is only 16,569 base pairs long and contains only 37 genes. Every human cell contains between 100 and 10,000 copies of mitochondrial DNA.
Unlike nuclear DNA, mitochondrial DNA does not recombine and thus there is no change between parent and child. Most importantly, mtDNA is only passed on from mother to child; although males inherit mtDNA from their mothers, they do not pass it on to their children. This unique feature of mtDNA allows it to be used for tracing matrilineage, the inheritance of mtDNA from mother to child.
When mtDNA is tested for genealogical purposes, a region of the DNA is sequenced for SNP mutations. mtDNA is divided into three regions “the coding region and two hyper-variable regions (HVR1 and HVR2). Most companies sequence the HVR1 (16001-16569), with HVR2 (073-577) and full-length (1-16569) sequencing becoming more and more useful for comparison. The DNA sequence is then compared to a single (randomly-chose) mtDNA sequence, the Cambridge Reference Sequence. The differences are listed as mutations that can be compared to the thousands of other mtDNA mutation lists that are stored in publicly-available databases. The results can also be used to determine the amount of time in which two individuals shared a most recent common ancestor (MRCA). The results of the mtDNA test can also be used to determine a person’s mtDNA haplogroup and haplotype.
Autosomal:
Autosomal DNA is the 22 pairs of non-sex chromosomes found within the nucleus of every cell. Autosomal DNA tests examine SNPs, or alleles, located throughout all of the DNA.
One of the most popular uses of autosomal DNA testing is to determine an individual's ethnic heritage. According to recent research, ethnic groups can contain distinctive alleles that are different from all other ethnic groups. The presence of that allele in an individual's DNA suggests that they are descended from that ethnic group. It should be noted, however, that the accuracy of these tests are still highly debated among scientists.
Some companies disclose the results as what percentage of each ethnic group the individual is, such as Native American, European, East Asian, and African. Other tests only look for a certain class of markers to reveal a certain type of ancestry, such as Native American, distinct African groups, Cohanim, Hindu, or European.
Genetic genealogy is another tool for the genealogist's toolbox. It is the use of genetics to study genealogy, the relationship between individuals.
There are at least four types of genealogical DNA testing, including Y-chromosome, X-chromosome, mtDNA, and autosomal DNA test.
Y-chromosome:
The Y chromosome is only found in males, who have one Y chromosome and one X chromosome. The Y chromosome is composed of 58 million base pairs and contains 83 genes which code for only 23 proteins.
During meiosis (preparation of cells for reproductive purposes) the Y chromosome and the X chromosome only recombine at the very ends of the chromosomes (roughly 5%) in a region called the telomeres. Most of the Y chromosomes are located in this region, meaning that they are shared between both sex chromosomes.
The lack of recombination means that the Y chromosome is passed down from father to son without significant change. Over long periods of time the chromosome begins to accumulate mutations that are typically silent and have no impact on the carrier. These mutations, however, are useful for genealogical purposes “they can be used to analyze the relationships between populations and individuals.
When a Y chromosome is submitted for analysis, short segments of DNA known as short tandem repeats (STRs) are sequenced. STRs are also known by their DYS number (DNA Y-chromosome Segment number). The number of repeats can change over time and are passed on from father and son, giving scientists the ability to trace paternal lineages. STR analysis typically provides a person's haplotype, although they are often used to suggest which haplogroup an individual matches.
Some genetics tests also analyze SNPs (Single Nucleotide Polymorphisms), single nucleotide changes in the DNA sequence. SNPs are typically identified using enzymes that cleave DNA – a change in the DNA sequence can prevent the enzyme from cleaving the DNA, creating a pattern that can easily be analyzed. SNP analysis typically provides a person's haplogroup.
Most Y chromosome tests examine between 12 and 67 STR markers, but many more are known and could eventually be used for relationship analysis. Each STR has a numerical value. DYS392, for example, has a value of 11, 12, 13, 14, 15, of 16, with 13 being the most common. The numerical values for all the STRs tested can then be compared to the thousands of other numerical sets available in DNA databases. If two people have identical numerical values, it is more likely that they have a recent common ancestor. Statistical analysis can be used to calculate the number of generations to the most common recent ancestor (MRCA).
X-chromosome:
The X chromosome, found in both males and females, is more than 153 million base pairs and contains roughly 1000 genes. Females have two X chromosomes while males have just one.
The use of X chromosomes to study genealogical relationships is still relatively new. The X chromosome, just like the Y, contains STRs, called X-STRs. The problem with studying X-STRs is that the entire X chromosome undergoes recombination during meiosis. In other words, in females the two X chromosomes randomly swap information and genes. Family Tree DNA, one of the leaders of X-STR research (using methods developed by DNA-Fingerprint), uses “haplotype blocks, or regions of X-STRs that are inherited intact over several generations.
A male's X chromosome is inherited from his mother and is a mixture of her two X chromosomes, one from her mother and one from her father. It is therefore a mixture of the maternal grandparent's X chromosomes.
A female inherits one X chromosome from each of her two parents. The X chromosome from her father is passed on from his mother is a mixture of her parents (the paternal great-grandparent's) DNA, while the X chromosome from the female's mother is a mixture of her parent’s (the maternal grandparent's) DNA.
mtDNA:
Mitochondrial DNA (mtDNA) is a small circle of DNA that is located inside a small organelle found inside our cells, the mitochondria. mtDNA is only 16,569 base pairs long and contains only 37 genes. Every human cell contains between 100 and 10,000 copies of mitochondrial DNA.
Unlike nuclear DNA, mitochondrial DNA does not recombine and thus there is no change between parent and child. Most importantly, mtDNA is only passed on from mother to child; although males inherit mtDNA from their mothers, they do not pass it on to their children. This unique feature of mtDNA allows it to be used for tracing matrilineage, the inheritance of mtDNA from mother to child.
When mtDNA is tested for genealogical purposes, a region of the DNA is sequenced for SNP mutations. mtDNA is divided into three regions “the coding region and two hyper-variable regions (HVR1 and HVR2). Most companies sequence the HVR1 (16001-16569), with HVR2 (073-577) and full-length (1-16569) sequencing becoming more and more useful for comparison. The DNA sequence is then compared to a single (randomly-chose) mtDNA sequence, the Cambridge Reference Sequence. The differences are listed as mutations that can be compared to the thousands of other mtDNA mutation lists that are stored in publicly-available databases. The results can also be used to determine the amount of time in which two individuals shared a most recent common ancestor (MRCA). The results of the mtDNA test can also be used to determine a person’s mtDNA haplogroup and haplotype.
Autosomal:
Autosomal DNA is the 22 pairs of non-sex chromosomes found within the nucleus of every cell. Autosomal DNA tests examine SNPs, or alleles, located throughout all of the DNA.
One of the most popular uses of autosomal DNA testing is to determine an individual's ethnic heritage. According to recent research, ethnic groups can contain distinctive alleles that are different from all other ethnic groups. The presence of that allele in an individual's DNA suggests that they are descended from that ethnic group. It should be noted, however, that the accuracy of these tests are still highly debated among scientists.
Some companies disclose the results as what percentage of each ethnic group the individual is, such as Native American, European, East Asian, and African. Other tests only look for a certain class of markers to reveal a certain type of ancestry, such as Native American, distinct African groups, Cohanim, Hindu, or European.
|
|
|
The history and geography of human genes 1994
By Luigi Luca Cavalli-Sforza, Paolo Menozzi, Alberto Piazza
http://books.google.com/books?id=FrwNcwKaUKoC&pg=PA493&lpg=PA493&dq=new+system+of+Q+haplotypes&source=bl&ots=Hl3UQfFFb4&sig=jm8qZTMHuYbTU-SWdebKaVFxtyA&hl=en&ei=FQ2gTcfuFIHeiAKL8YyKAw&sa=X&oi=book_result&ct=result&resnum=10&ved=0CEUQ6AEwCQ#v=onepage&q=new%20system%20of%20Q%20haplotypes&f=false
By Luigi Luca Cavalli-Sforza, Paolo Menozzi, Alberto Piazza
http://books.google.com/books?id=FrwNcwKaUKoC&pg=PA493&lpg=PA493&dq=new+system+of+Q+haplotypes&source=bl&ots=Hl3UQfFFb4&sig=jm8qZTMHuYbTU-SWdebKaVFxtyA&hl=en&ei=FQ2gTcfuFIHeiAKL8YyKAw&sa=X&oi=book_result&ct=result&resnum=10&ved=0CEUQ6AEwCQ#v=onepage&q=new%20system%20of%20Q%20haplotypes&f=false
EthnoAncestry is an innovative DNA testing company formed in 2004
with the aim of bringing the cutting edge of genetic genealogy research
to people through the development of new genetic signatures and
markers, and by providing authoritative interpretation of deep ancestry.
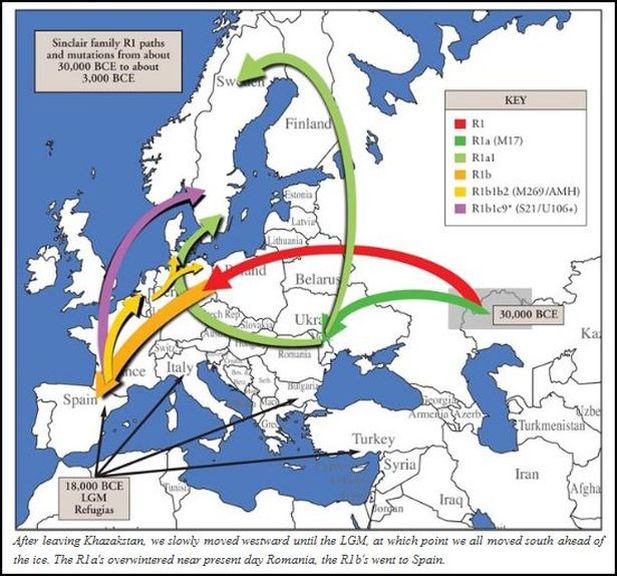
This is what Alistair Moffatt and Jim Wilson's book has to say about L21 people. The book doesn't mention the L193 SNP, but this is the group it's part of: "Toward the end of the third millennium BC, the genetic make-up of Scotland was beginning to divide. A British variant of the European marker, M269, known as S145 [or L21] could be said to be the most emphatic signal of the Celtic language speakers of the British Isles. It is found in England, Wales, Scotland and Ireland and it is almost certainly characteristic of those farming communities who may have spoken early forms of Celtic languages in the centuries around 2,000 BC."
"DNA sampling reinforces an intertwined sense of two distinct seaborne trading networks in Britain and Ireland. In the west, the emphatic presence of S145 appears to mirror mercantile contact. Distinctive pots known as maritime bell beakers were first made in the region around the River Tagus in Portugal and the tradition of bows and arrows in graves may also have originated from there. By 2,500 BC this cultural package had spread north to the Morbihan area of southern Brittany and the mouth of the Loire. This area became a centre of production and exchange not only for bell beakers but also other valuable items such as axes, flints, daggers and lance heads. From Morbihan/Loire, the beakers filtered down the French river valleys to the Mediterranean coast and eastwards towards northern Italy. To the north, contacts were made with Wessex, Ireland and Atlantic Scotland. Now, it appears that S145 also travelled these trading routes. The marker probably originated in southern France and northern Iberia and people carrying it came to Ireland and western Scotland. This was not a wave of immigration, but a series of small movements over time, probably in the millennium between 2,500 BC and 1,500 BC."
http://scotsfamily.com/dna-testing.htm
Bursting with surprises, the DNA studies track migration over millennia, primarily since the end of the last Ice Age, around 12,000 years ago. As ice receded, humans returned to the Isles over land currently under the North Sea and the English Channel, when the Thames was a tributary of the Rhine. As the warm-up progressed, the seas rose, creating the forested Isles. The professor warns, We are now in a warm phase of the long-term glacial cycle, but it will not last forever, and at some as yet unpredictable time in the future we will slide inexorably into another Ice Age.
Readers trudge alongside Sykes and his crew from the Oxford Genetic Atlas Project into rural classrooms, blood donor sites, and local festivals in search of DNA. The author states that, It took a lot of mental effort constantly to remind myself that every single one of these strings of letters and numbers represented the journey of an ancestor. What combos of indigenous Britons (Celts), Romans, Scottish Celts and Picts, Irish Celts, Welsh Celts, Vikings, Saxons and Anglo-Normans are stirring within the genetic soup of the Isles today? Matchmaker Sykes introduces myth to scientific methodology when answering these questions.
England, the most crowded region in the Isles today, was invaded for a thousand years beginning with the Romans in 43 AD. Asserting that tales of 6th century King Arthur and the ancient Britons are rooted in fact, the professor remarks that, In my research around the world I have more than once found that oral myths are closer to the genetic conclusions than the often ambiguous scientific evidence of archaeology. Arthurian traditions faded when Henry VIII broke with the Catholic Church in the 16th century, and the resulting veneration of Saxon King Alfred provoked centuries of Saxon vs. Celt conflict. This conjuring of a new origin myth peaked in the 19th century with Saxon superiority characterized by a righteous, crusty citizenry towering over low-life Celtic loungers. The Roman Empire’s collapse offered expansion opportunities to barbarian Angles, Saxons and Jutes from the 8th to the 11th centuries, and the DNA of these tough customers remains strong in northeastern England. The bloodlines of William of Normandy, leader of the Norman Invasion in 1066, render him a recycled Viking.
Saxon, Dane and Norman are close German/Scandinavian cousins; our tongue is English instead of Celtic, courtesy of these invaders. In addition to donations of Viking, Saxon and Norman blood, a smattering of African and Middle Eastern DNA was found in southern England, shocking individuals with no such known ancestry. These DNA dribbles lead the author to think that these folks may be descended from Roman slaves. History lessons keep surging out of this grand field trip, and they alone are worth the price of the book. The professors instruction on Irish Celts is absorbing. Although the Vikings elbowed their way around Ireland during the 9th century, Celtic genetic dominance is huge. Folks in Leinster show some Anglo-Norman DNA influence, and an intriguing Y-chromosome ancestral study of males with old Gaelic names (Mc, Mac, O) had me in thrall.
Research in Scotland turned up Picts, Celts from Ireland, Vikings and Anglo-Normans. Most Scots are genetically similar to the Irish, amazingly so in Argyll. Old camps show Mesolithic people in Scotland at Orkney and the Shetlands 10,000 years ago, and the exception to the genetic closeness with Ireland rests in these areas. Vikings began arriving in the neighborhood during the late 8th century, and Norse place names still dominate the landscape. DNA studies prove that the occupiers brought their own women along, and Viking ancestry stands at 30 to 40 percent today. Sykes describes a wee Scandinavian air in Shetland as an “Undemonstrative, no-nonsense feeling of the place.” I have always wondered about the Picts – Celtic variants or a relic population? The Romans called them Picti (Painted People). This treasure hunt demands reading in one sitting. Similarities with Celts were found, and DNA testifies that Picts flourish among us still, predominately in the Grampian and Tayside regions of Scotland.
Wales withstood assaults by Romans, Irish, Saxons, Normans, and English, and it manifests a mighty genetic kinship with Ireland and Scotland, minus the Viking donors. Some individuals living in the remote mountains near Plynlimmon and Tregaron found themselves the focus of much early 20th century research on their odd-shaped heads and Neanderthal-like faces. One extraordinary anecdote concerns two brothers who were widely regarded as Neanderthals and notoriously named the Tregaron Neanderthals. In the 1950s and 1960s, local Welsh teachers, following instructions in their schools history syllabi, took students to interview these welcoming men. The brothers died in the 1980s, and Sykes doubts that they were true Neanderthals, but scientists are still looking out for Neanderthal DNA (none found to date).
The professor’s reports confirm that almost everyone in Ireland and Britain has Celtic ancestors who arrived thousands of years ago in flimsy boats over the Atlantic from Celtic Iberia. These findings give credence to the Irish Milesian myth, and Sykes reiterates that,”Deeply held origin myths, however richly embroidered, have a habit of being right.” This book remains unforgettable as Bryan Sykes reminds that,”within each and every one of our cells is something that has witnessed every life we have ever lived.” Laced with fact and folklore from an ancient heritage and rich with lashings of comedy throughout DNA collection capers, SAXONS, VIKINGS, AND CELTS dazzles as blood and bone stand up and testify.
The Picts are believed to be the descendants of the first people to colonise Scotland after the glaciers retreated at the end of the last ice age. Until recently the degree to which present day Scots (and their diaspora) are descended from these mysterious people was unknown. However, recent genetic analysis has revealed the existence of two Pictish Y chromosome signatures which are most common in Scotland, but rarely seen in England or continental Europe.
This is what Alistair Moffatt and Jim Wilson's book has to say about L21 people. The book doesn't mention the L193 SNP, but this is the group it's part of: "Toward the end of the third millennium BC, the genetic make-up of Scotland was beginning to divide. A British variant of the European marker, M269, known as S145 [or L21] could be said to be the most emphatic signal of the Celtic language speakers of the British Isles. It is found in England, Wales, Scotland and Ireland and it is almost certainly characteristic of those farming communities who may have spoken early forms of Celtic languages in the centuries around 2,000 BC."
"DNA sampling reinforces an intertwined sense of two distinct seaborne trading networks in Britain and Ireland. In the west, the emphatic presence of S145 appears to mirror mercantile contact. Distinctive pots known as maritime bell beakers were first made in the region around the River Tagus in Portugal and the tradition of bows and arrows in graves may also have originated from there. By 2,500 BC this cultural package had spread north to the Morbihan area of southern Brittany and the mouth of the Loire. This area became a centre of production and exchange not only for bell beakers but also other valuable items such as axes, flints, daggers and lance heads. From Morbihan/Loire, the beakers filtered down the French river valleys to the Mediterranean coast and eastwards towards northern Italy. To the north, contacts were made with Wessex, Ireland and Atlantic Scotland. Now, it appears that S145 also travelled these trading routes. The marker probably originated in southern France and northern Iberia and people carrying it came to Ireland and western Scotland. This was not a wave of immigration, but a series of small movements over time, probably in the millennium between 2,500 BC and 1,500 BC."
http://scotsfamily.com/dna-testing.htm
Bursting with surprises, the DNA studies track migration over millennia, primarily since the end of the last Ice Age, around 12,000 years ago. As ice receded, humans returned to the Isles over land currently under the North Sea and the English Channel, when the Thames was a tributary of the Rhine. As the warm-up progressed, the seas rose, creating the forested Isles. The professor warns, We are now in a warm phase of the long-term glacial cycle, but it will not last forever, and at some as yet unpredictable time in the future we will slide inexorably into another Ice Age.
Readers trudge alongside Sykes and his crew from the Oxford Genetic Atlas Project into rural classrooms, blood donor sites, and local festivals in search of DNA. The author states that, It took a lot of mental effort constantly to remind myself that every single one of these strings of letters and numbers represented the journey of an ancestor. What combos of indigenous Britons (Celts), Romans, Scottish Celts and Picts, Irish Celts, Welsh Celts, Vikings, Saxons and Anglo-Normans are stirring within the genetic soup of the Isles today? Matchmaker Sykes introduces myth to scientific methodology when answering these questions.
England, the most crowded region in the Isles today, was invaded for a thousand years beginning with the Romans in 43 AD. Asserting that tales of 6th century King Arthur and the ancient Britons are rooted in fact, the professor remarks that, In my research around the world I have more than once found that oral myths are closer to the genetic conclusions than the often ambiguous scientific evidence of archaeology. Arthurian traditions faded when Henry VIII broke with the Catholic Church in the 16th century, and the resulting veneration of Saxon King Alfred provoked centuries of Saxon vs. Celt conflict. This conjuring of a new origin myth peaked in the 19th century with Saxon superiority characterized by a righteous, crusty citizenry towering over low-life Celtic loungers. The Roman Empire’s collapse offered expansion opportunities to barbarian Angles, Saxons and Jutes from the 8th to the 11th centuries, and the DNA of these tough customers remains strong in northeastern England. The bloodlines of William of Normandy, leader of the Norman Invasion in 1066, render him a recycled Viking.
Saxon, Dane and Norman are close German/Scandinavian cousins; our tongue is English instead of Celtic, courtesy of these invaders. In addition to donations of Viking, Saxon and Norman blood, a smattering of African and Middle Eastern DNA was found in southern England, shocking individuals with no such known ancestry. These DNA dribbles lead the author to think that these folks may be descended from Roman slaves. History lessons keep surging out of this grand field trip, and they alone are worth the price of the book. The professors instruction on Irish Celts is absorbing. Although the Vikings elbowed their way around Ireland during the 9th century, Celtic genetic dominance is huge. Folks in Leinster show some Anglo-Norman DNA influence, and an intriguing Y-chromosome ancestral study of males with old Gaelic names (Mc, Mac, O) had me in thrall.
Research in Scotland turned up Picts, Celts from Ireland, Vikings and Anglo-Normans. Most Scots are genetically similar to the Irish, amazingly so in Argyll. Old camps show Mesolithic people in Scotland at Orkney and the Shetlands 10,000 years ago, and the exception to the genetic closeness with Ireland rests in these areas. Vikings began arriving in the neighborhood during the late 8th century, and Norse place names still dominate the landscape. DNA studies prove that the occupiers brought their own women along, and Viking ancestry stands at 30 to 40 percent today. Sykes describes a wee Scandinavian air in Shetland as an “Undemonstrative, no-nonsense feeling of the place.” I have always wondered about the Picts – Celtic variants or a relic population? The Romans called them Picti (Painted People). This treasure hunt demands reading in one sitting. Similarities with Celts were found, and DNA testifies that Picts flourish among us still, predominately in the Grampian and Tayside regions of Scotland.
Wales withstood assaults by Romans, Irish, Saxons, Normans, and English, and it manifests a mighty genetic kinship with Ireland and Scotland, minus the Viking donors. Some individuals living in the remote mountains near Plynlimmon and Tregaron found themselves the focus of much early 20th century research on their odd-shaped heads and Neanderthal-like faces. One extraordinary anecdote concerns two brothers who were widely regarded as Neanderthals and notoriously named the Tregaron Neanderthals. In the 1950s and 1960s, local Welsh teachers, following instructions in their schools history syllabi, took students to interview these welcoming men. The brothers died in the 1980s, and Sykes doubts that they were true Neanderthals, but scientists are still looking out for Neanderthal DNA (none found to date).
The professor’s reports confirm that almost everyone in Ireland and Britain has Celtic ancestors who arrived thousands of years ago in flimsy boats over the Atlantic from Celtic Iberia. These findings give credence to the Irish Milesian myth, and Sykes reiterates that,”Deeply held origin myths, however richly embroidered, have a habit of being right.” This book remains unforgettable as Bryan Sykes reminds that,”within each and every one of our cells is something that has witnessed every life we have ever lived.” Laced with fact and folklore from an ancient heritage and rich with lashings of comedy throughout DNA collection capers, SAXONS, VIKINGS, AND CELTS dazzles as blood and bone stand up and testify.
The Picts are believed to be the descendants of the first people to colonise Scotland after the glaciers retreated at the end of the last ice age. Until recently the degree to which present day Scots (and their diaspora) are descended from these mysterious people was unknown. However, recent genetic analysis has revealed the existence of two Pictish Y chromosome signatures which are most common in Scotland, but rarely seen in England or continental Europe.
"The
probability that human intelligence developed all the way from the
chemical ooze of the primeval ocean solely through random sequences of
random mechanical processes has been aptly compared to the probability
of a tornado blowing through a gigantic junkyard and assembling by
accident a 747 jumbo jet." - Stanislov Grof
GENETIC MEMORY
THE DNA spiral embodies the long and winding road of human history....
THE DNA spiral embodies the long and winding road of human history....
The structure of DNA as we know it is made up of letters and thus has a
specific text and language. You could say our bodies are made up of
language, yet we assume that speech arises from the mind. How do we
access this hidden language? By studying it. There are several roads to knowledge, including science and shamanism. The
symbol of the Cosmic Serpent, the snake, is a central theme in your
story, and in your research you discover that the snake forms a major
part of the symbology across most of the world’s traditions and
religions. There is a consistent system of natural symbols in
the world.
|
|
|
A genetic marker is a variation in the nucleotide sequence of the
DNA, known as a mutation. Mutations which occur within genes—a part of
the DNA which codes for a protein—usually cause a malfunction or disease
and is lost due to selection in succeeding generations. However,
mutations found in so-called “non-coding regions” of the DNA tend to
persist. Since the Y chromosome consists almost entirely of non-coding DNA
(except for the genes determining maleness), it would tend to accumulate
mutations. Since it is passed from father to son without recombination,
the genetic information on a Y chromosome of a man living today is
basically the same as that of his ancient male ancestors, except for the
rare mutations that occur along the hereditary line. A combination of these neutral mutations, known as a haplotype, can
serve as a genetic signature of a man’s male ancestry. Maternal
genealogies are also being studied by means of the m-DNA (mitrocondrial
DNA), which is inherited only from the mother.
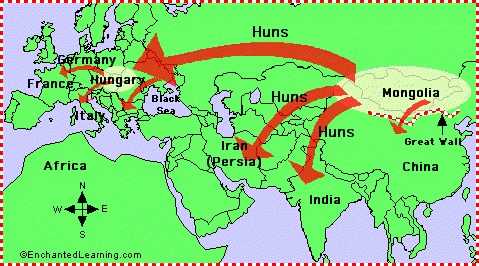
The Huns (Xiongnu/Huns)
----------------------------------------
The Hun tribes, or as the Chinese called them the Xiongnu, stemmed basically from the Siberian
branch of the Mongolian race.
http://www.silk-road.com/artl/xiongnu1.shtml
The Xiongu > The Huns
------------------------------------
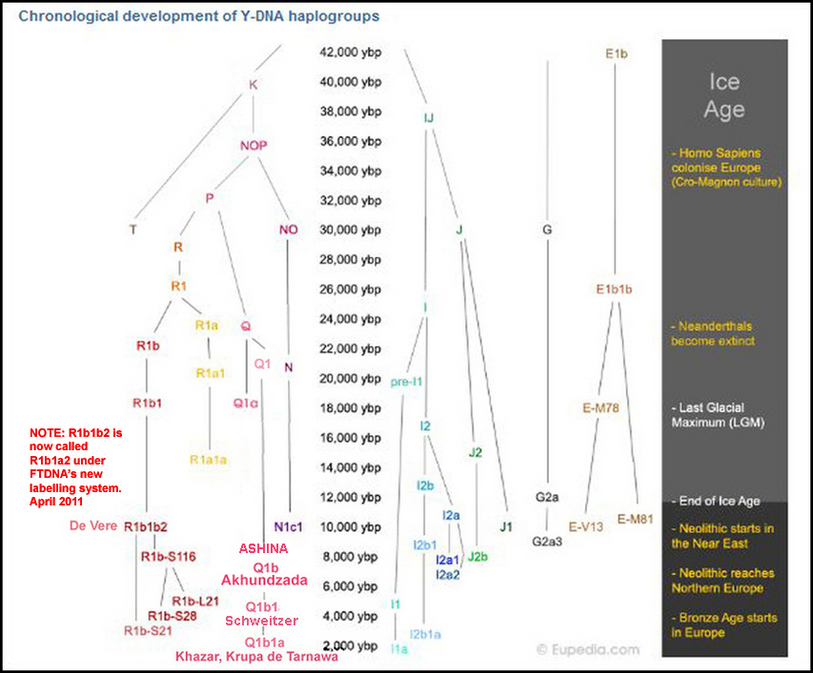
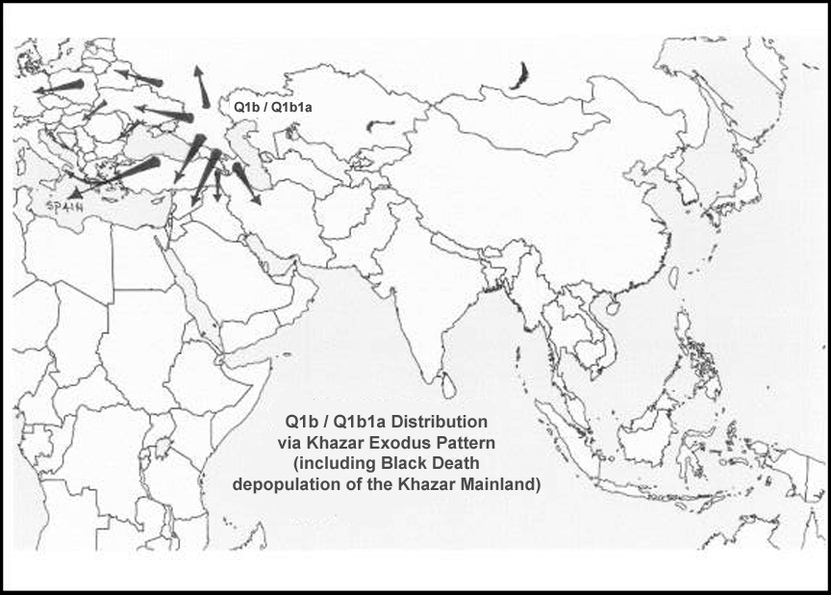
Despite the fact that Mongolia is still one of the most isolated and one of the most sparsely populated countries in world, Y-DNA Q1b (haplogroup very rare in human populations for itself) has been detected by researchers from Department of Legal Medicine and Bioethics, Graduate School of Medicine, Nagoya University, Japan in all five regional populations (Ulaangom, Dalandzadgad, Ulaanbaatar,Undurkhaan,Choibalsan) in Mongolia and even in Japanese population (日本人 Nihonjin, Nipponjin).
Therefore this finding is of etreme importance due to the reason that near Mongolian southern border is location of cemetery Heigouliang, Xinjiang (Black Gouliang cemetery), east of Barkol basin, near the city of Hami. By typing results of DNA samples during the excavation of one of the tombs it was determined that of the 12 men there were: Q1a - 6, Q1b (M378) - 4, Q-2 (unable to determine subclade). Hosts of the tombs were representatives of Y-haplogroup Q1b-M378 exclusively (while Y-DNA Q1a represents sacrificial victims). They date from the time of early (Western) Han (2-1 century BC). In the very same location today Turkic Uygurs shows Q1b haplotypes very close if not the same as Ashkenazi Jewish Q1b(1a).
Source of attached table: http://www.fsigeneticssup.com/…/S1875-1768%2813%2…/fulltext…
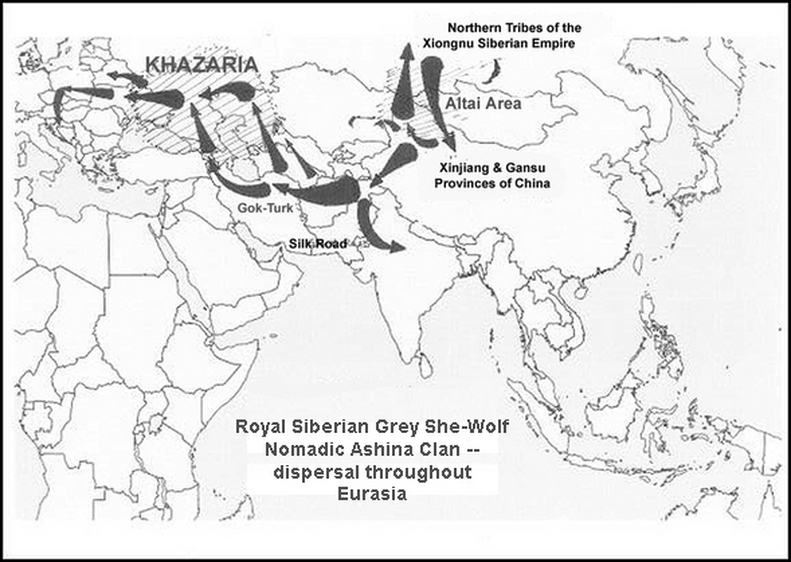
Ashina was a branch of Xiongnu.
The Xiongnu established two dynasties. Both dynasties were organized as nomads. Among the united tribes there was
supposedly a tribe called Ashina (which was the name of the then leading clan of the steppe nomads or Turk (Turkut).
It's stated that the Ashina were related to the Northern branch of the Xiongnu.
(According to the New Book of Tang, the Ashina were related to the northern tribes of the Xiongnu, in particular they were of Tiele tribe by ancestral lineage.)
(See the part, below, on Hun Dynastic mating practices.)
It's further stated that the Northern branch of the Xiongnu, left the federation and become known as the Huns.
The Huns are Q1a2 and Q1b
------------------------------------------
This material is taken directly from this site, with limited paraphrasing:
Quotes:
Other Terms for Huns
---------------------------------
In the earlier pre-historic period the Huns were called Hu.
In the late pre-historic period the Huns were called Hun-yui; during the Zhou period (1045–256 BC) they were called Hyan-yun; starting from the Qin period (221-206 BC) they were called Hunnu.
The Tele Tribes
------------------------
The Tele (Tiele) were a branch of the Huns.
The Tele tribes emerged after the disintegration of the Xiongnu confederacy.
The Tele left the early Huns Horde, keeping patriarchal relations and a nomadic life.
The Tele were a collection of tribes of different Turkic ethnic origins.
The leaders of the Hun tribe had a greater level of mongoloidness than that of the members of the Tele (estimated from biological studies, as reaching 89%).
http://webcache.googleusercontent.com/search…
Tele Tribes and Uigurs
-----------------------------------
Besides that, the known ancient written sources indicate the origin of some Turkic-speaking tribes (belonging to the Tele group) can be traced directly from the Huns. Take the Uigurs...Tangshu directly tells that Uigur ancestors were Huns. In the earlier dynastic history, the ancestors of the Tele tribes, in particular the Uigurs, are derived from the Huns.
(Note: This fits with the Hun Dynastic model.See the Hun Dynastic Model.)
The nearest historical ancestors of modern Southern Altaians were mainly Tele tribes, and four centuries ago coached in the territory of Siberia, not only in its southern, mountain part in the Sayano-Altai mountains, but also in
the forest-steppes.
The so-called "real Huns" during the Ancient Turkic time have not disappeared at all. A number of tribes from the ethnic composition of the Huns have joined, probably partially, under their tribal names, partially under changed names, in the confederation of the Tele tribes, or in the Ancient Turkic Kaganates. Among the Tele tribes, one tribe even had the folk name of Hunnu (Huns) and it's known that it was called Hun.
Mongoloid Descent Noted
----------------------------------------
Turkic So, was stated to be the primogenitor tribe of Turk tribes.
The Turkic Tele Uigur clan, Toba, was also stated to be the founder dynastic clan. Toba descended from the tribe/country So (So-lu), which, as was stated, was the progenitor ancestor of the Turks.
Toba were Tungus Tanguts = future Mongol Uhuans (Wuhuan).
The Dulo was the Tele Tribes
--------------------------------------------
The Tokuz-Oguzes are called "nine tribes", which is a translation into the Chinese language of the term Tokuz-Oguz. The name Tokuz-Oguz means a confederation of the 9 Tele tribes.
In Middle Asia "Tele" was pronounced as "Dulo", and in that form it appears in the dynastic and historical records.
The Tele Kaganate was headed by Seyanto leaders. Uigurs also played a role.
The political influence of the (Tele) Kaganate headed by Seyanto extended to the territory of the modern Mongolia, Tuva, Altai, and the Hyagas state, located in the Minusinsk depression.
At the end of the 6th century the Western Turks controlled the lands north from the Gobi, and a majority of the Tele tribes fell under their rule.
Conflict between the Ashina/China and
-----------------------------------------------------------
Seyanto (of the Tele tribes)
-----------------------------------------
The tribal aristocracy of the eastern Turks, subordinated to China, elected as a leader Kupi, from the Kagan clan Ashina.
In 632 to 635 Teles crushed the Western Turkic Kagans. The Tokuz-Oguzes were led by Seyanto.
Kupi established his court to the north of the Mongolian Altai, where he proclaimed himself Turkic Kagan.
In 646 the Chinese army, together with the calvary of the eastern Turks trounced Seyanto. Seyanto was completely broken and dispersed.
http://s155239215.onlinehome.us/…/201Alt…/Potapov-TeleEn.htm
The Tele and the Ashina
-------------------------------------
There appear to be no written records that list the Ashina and Dulo as being listed as being one of the 12 or 15 tribes of the Tele Tribes.
Although there is this...
Quote
Bichurin 1.4 recites 12 Tele ancestral tribes in Ch. rendition see analysis by Gumilev:
The first two...
1, Lifuli (??????) (Barsil? "fu-li" is "bori"
= Tr. wolf)
2. Tulu (????) (Tunlo? Dulo? Dubo?)
However, there are also references that associate the Ashina with with Tele tribes.
Note that the Dulo and the Ashina usedthe same tagma symbol.
However, the Ashina were not actually listed as being one of the 12 or 15 tribes of the Tele tribe. Therefore one has to interpret the term "...they were of the Tiele tribe by ancestral lineage."
(See the part on Hun Dynastic mating
practices.)
The Hun's Dynastic Clan
--------------------------------------
The Hun's dynastic clan was Uiguro-Hunnic, or Tele-Hunnic. They were the Hun tribe's papas and Tele/Uigur tribe's mamas. The tribes of Huns and Tele/Uigurs were permanent mutual conjugal partners, a custom retained among the Turkic peoples up to the present time, and a major tool in a scientific research.
Hun tribe's Papas + Tele/Uigur tribe's Mamas
The Hun/Uigur conjugal union ascends to the time before their appearance in the Chinese annals, and the looks of the dynastic Huns and Uigurs was sufficiently blended after centuries, if not a millennium, of confined cross-breeding.
http://webcache.googleusercontent.com/search…
Notes:
Uyghur/Uigur--Originally known as the Dingling, an ancient Siberian people.
The Uigur's ancestors were Huns:
Uigur ancestors were Huns. In the earlier dynastic history, the ancestors of the Tele tribes (The Dulo), in particular the Uigurs, are derived from the Huns.
(Note: This fits with the Hun dynastic model.)
The Ashina were Huns
-----------------------------------
Among the united tribes, of the Xiongnu federation, there was a tribe called Ashina, which was the name of the leading clan of the steppe nomads (Turk).
From the dynastic history, it was already known that the ancestors of the ancient Turks, under the name Ashina, were a separate branch of the house of Hunnu (Huns).
The ethnogenetical connection of the ancient Turks with the Hunnu (Huns) was definitely stated by the source.
The ancestors of the ancient Turks, under a name Ashina, were a separate branch of the house of Hunnu (Huns)."
-- the Zhou (551–583) dynastic history
The Ashina were actually Huns, and Iranian
------------------------------------------------------------------
Barbarians
-----------------
According to the New Book of Tang, the Ashina were related to the northern tribes of the Xiongnu, in particular they were of the Tele tribe by ancestral lineage.
(See the part on Hun Dynastic mating practices.)
According to the Book of Zhou and History of Northern Dynasties, the Ashina were a branch of the Xiongnu, while according to the Book of Sui and Tongdian they were "a mixture of Hu" from Pingliang.
(Note: Hu means Huns.)
(See the part on Hun Dynastic mating
practices.)
There is no doubt that the Turks came from a mixing of the late Huns, who penetrated to the west after 265 AD (during a mass migration to the west of the Hunnic tribes from Central Asia), in the area of the small Pinlyan and Hesi states, with the local Iranian-lingual "barbarians."
Hun Tribes
The Ashina Turk type of the Huns descended from the Hun tribe Se/So.
Ashina was a "member tribe" of the So/Se/Sek/Saka tribes.
So/Se/Sek/Saka was the Parental tribe of the Ancient Turks.
The Saka were a Scythian tribe or group of tribes of Iranian origin.
Turkic So, was stated to be the primogenitor tribe of Turk tribes.
From there, after a defeat of Pinlyan by China in the 2nd half of the 5 century, the ancestors of the Turks went to the Gaochan mountains, and were subjugated by Jujans, and then settled by Jujans on the southern slopes of Mongolian Altai, where they were engaged in metalwork for the Jujan kagans.
Notes:
The migration of the Turks occurred under the Asina clan (originally Hiung-nu, so Asiatic Hun).
More accurately, it's stated, the Hunnic tribe Saka [Saka/Se] is given by Chinese annals of the later Wei Dynasty.
http://webcache.googleusercontent.com/search?
q=cache:3cbOa1xTU4wJ:s155239215.onlinehome.
us/turkic/23Avars/InMemoryOfAvarKhagans.htm+
the+Hun+tribes+th+so+clan&cd=16&hl=en&ct=
clnk&gl=us
This makes sense in light of other information on the Ashina: It's stated that the Ashina Turk type of the Huns descended from the Hun tribe Se/So. Some authors have implied that there is an Iranian link between the Se/So and the
Ashina.
It's also stated that theTurks descended from the tribe So that lived in the northern Altai.
Hun's tribes--Dominating Turkic dynastic tribe Sary (Se).
Se > Chinese term.
Se = Sakas
It's stated that the Se-yanto or Syr-Tardush were an ancient Tele people.
Seyanto (Sir + Yanto)--The Syr-Tardush
In 631, the Seyanto state successfully controlled all Turk's leaders except for Ordos Turks under Chinese protection.
It's also stated that Seyanto and Bugu had identical tamgas. (Note so did the Ashina and the Dulo...? However, also note that the Tele was known as the Dulo in central Asia.)
Notes:
Although the Tele was also known as the Dulo, different sources also make distinctions:
The On Okh (Onoq, Ten Arrows) or Western Turks--Subdivision: The greater Tardush (Tulu, Dulo) <----
---Branch: Turgesh
The Turgesh were a Turkic tribal confederation who emerged from the ruins of the Western
Turkic Kaganate.
This indicates that the greater Tardush was the Tulu, Dulo clan.
http://en.wikipedia.org/wiki/Turgesh
The Turgesh were a group of Duolu tribes .
The Foundation of the Turgesh Kaganate was Precipitated by Anti- Ashina Turgesh rebellion. Ashina--The Counter-Movement of Turgeshes, ended with a capture of six hundred and ninety-nine in Suyab .
http://translate.google.com/translate…
Notes:
The Dulo and the Ashina were also referred to as being cousins, in other sources, who became opposed to each other, as the result of ethnic tensions.
The Khazar kingdom was ruled by the Ashina dynasty of Western Turks (Turkuts), who took over the territory.
The On Okh (Onoq, Ten Arrows) or Western Turks--Subdivision: The greater Tardush (Tulu, Dulo) <----
---Branch: Turgesh
http://www.jstor.org/discover/10.2307/41035790…
Comments
-----------------
The Xiongu > The Huns (Q1a2 and Q1b) > Ashina
(See the part on Hun Dynastic mating practices.)
It appears that there was both a Q1a2 and a Q1b genetic core in the Huns, and based upon the Hun dynastic practices of mating, it appears that the Pappas would continue to be Q1a2 and Q1b, and that the Mammas came from the Tele tribes.
Being related to the Tele tribe tells one nothing specific, about haplogroups, but the presence of Q is indicated through the Ashina being related to the Northern branch of the Xiongnu, who became the Huns.
The Ashina were a separate branch of the house of Hunnu (Huns).
The Ashina were related to the Northern Branch of the Xiongnu/Huns who became known as the Northern Huns.
According to the Book of Zhou and History of Northern Dynasties, the Ashina were a branch of the Xiongnu, while according to the Book of Sui and Tongdian they were "a mixture of Hu" (Huns) from Pingliang.
According to the New Book of Tang, the Ashina were related to the northern tribes of the Xiongnu, in particular they were of the Tele tribe by ancestral lineage.
The Tele also left the Huns, keeping patriarchal relations.
The custom (Hun dynastic mating practices) was retained among the Turkic peoples up to the present time. Therefore, this implies that Hun Y-DNA (Q1a2 and Q1b) was passed down, throughthe Ashina males, and that the Turkic peoples
retained this custom.
Research has indicated that the Northern Huns genetic core was Q1a2 and Q1b.
Huns (Y: Q1a2 and Q1b) > Tele (MtDNA)
According to the Book of Sui and Tongdian the Ashina were a mixture of Hu (Huns) from Pingliang.
There is no doubt that the Turks came from a mixing of the late Huns, in the area of the small Pinlyan and Hesi states, with the local Iranian-lingual "barbarians."
Certain authors have traced Se/So/Saka to Iranian sources. What appeared to be contradictory material makes sense in light of the Hun dynastic mating model, because it connects all of this material together.
The Ashina were a seperate branch of the House of Huns. The ancestors (Ashina) of the Turks were a branch of the Huns.
The Ashina were the result of the intermixing of the Huns with Iranian Barbarians.
The view widely spread in the world of Oriental studies is that that Huns were ancestors of the ancient Turks as was repeatedly certified by the ancient Chinese written sources.
From the dynastic history it's known that the ancestors of the ancient Turks, under a name Ashina, were a separate branch of the house of Hunnu (Huns). The ethnogenetical connection of the ancient Turks with the Huns was definitely
stated by the source. But there is other historical information that allows one to detail and refine this connection.
There is no doubt that they came from a mixing of the late Huns, in the area of the small Pinlyan and Hesi states, with the local Iranian-lingual barbarians. From there, after a defeat of Pinlyan by China, the ancestors of the Turks coached
away to the Gaochan mountains, northwest from Turfan, and were subjugated by Jujans, and then settled by Jujans on the southern slopes of Mongolian Altai, where they engaged in metalwork for the Jujan kagans.
http://webcache.googleusercontent.com/search…
(This particular example of intermixing, applied to the creation of the Ashina, would explain why one can trace the Ashina origins in two directions. In light of this material, it's not a contradiction, as it is missing pieces of a puzzlethat puts everything into perspective by making it fit together.)
If the Tele was the Dulo, as was claimed, then, based upon the Hun dynastic model, it wouldindicate that the Northern Huns would have been Q1a2 and Q1b...Hence, the Ashina would have been Q1a2 or Q1b...Also, Atiila the Hun
was associated with Dulo (Tele Tribes), and according to research, Atiila the Hun was believed to be either Q1a2 or Q1b. That would also fit with the Hun dynastic model.
Ashina Dynastic Partners
---------------------------------------
The female dynastic partner of the Ashina Turks, with a title to the country land and people, is a pin. Yuezhi tribe, which is a Chinese codeword for the Tokhars.
http://s155239215.onlinehome.us/tu…/29Huns/CsornaiHunsEn.htm
Tahia (Tokhars)
The Yuezhi may have been a Europoid people, as indicated by the portraits of their kings on the coins they struck following their exodus to Transoxiana (2nd-1st century BCE)
Comments
-----------------
It further states that the Turks continued this dynastic practice. Therefore, it appears that Pappa Hun Y-DNA would have to continue to be either Q1a2 or Q1b. It appears that the Ashina (Hun) Pappas dynastic practices eventually involved mating with Tokhar women instead of women from the Tele tribe. This would indicate that the male descendants of Ashina males would continue to be either Q1a2 or Q1b.
Based upon these patterns, it appears thatthe only other Y-DNA haplogroup, that could be introduced into the Ashina line, would bethrough the dynastic partners of the Ashinawomen, who were from the Barc tribe, and
that is only if they weren't also Q1a2 or Q1b.
(Note the Barc tribe/clan was stated to havebeen brother-in-law material for the Ashina women.)
http://www.jstor.org/discover/10.2307/41035790…
Huns (Y-DNA: Q1a2 and Q1b) > Tele (MtDNA)
Hun/Ashina (Y-DNA: Q1a2 and Q1b) > Tokhars (MtDNA)
The following factor needs to be factored into this:
Ashina Women > Barc Tribe Males (Y: ?)
Notes for it are:
The non-qaganal clan of the Barc/Warâz/Bolcân, or VarazAVarac/Barc, or Varaz/Warac/Barc, who were another western turk clan.
nekagansky clan Bartsch / Varaz / Bolchan ( Barc / Waraz / Bolcan ) - corresponding dynasty Ihshad / Running / Iillig (Ihsed / Beg / Yillig)
Comments
-----------------
The Huns were Q1a2 and Q1b.
Atilla the Hun was from the Dulo (Tele) and he was believed to be Q1a2 or Q1b.
The Huns dynastic model involved mating practices where the Huns were the Pappas (Y-DNA Q1a2 or Q1b)
and the Mammas were from the Tele/Uigurs (mtDNA).
The Ashina were the result of Hun Y-DNA + Iranian Barbarian mtDNA. The Ashina were also associated
with the Tele.
Other sources connect the Ashina with So/Se, which is connected to Iranian sources, and this starts
making sense, in light of the above information.
Tele Tribes
1 Uange (Uygurs)
2 Seyanto (Sir + Yanto)
3 Kibi (Kibir)
4 Dubo (Tubalar)(Dabo)(Tele)
5 Guligan (Kurykan)(Yakut)
6 Dolange (Telengits)
7 Bugu (Pugu)(Uygurs)
8 Bayegu (Baiyrku)(Uygurs)
9 Tunlo (Tongra)(Uygurs)
10 Hun
11 Sygye (Uygurs)
12 Husye
13 Higye
14 Adye(Eduz)
15 Baysi (Barsil)
http://s155239215.onlinehome.us/…/71_…/hun_dateline_1_En.htm
It's stated that theTurks descended from the tribe So that lived in the northern Altai.
In the Hun's tribes, it's been stated that the dominating Turkic dynastic tribe was Sary
(Se).
Se > Chinese term.
There are also clans in tribe Se.
--A clan of tribe Se (Sir/Sary).
In 300 AD, Sirs and Turks lived at Ordos.
http://s155239215.onlinehome.us/…/71_…/hun_dateline_1_En.htm
It's stated that the Se-yanto or Syr-Tardush were an ancient Tele people.
Seyanto (Sir + Yanto)--The Syr-Tardush
It's stated that Alat-tou belonged to a clan oftribe Se (Sir/Sary), who at some time wiped out Yanto leadership, and took over leadership of the Sary/Yanto confederation.
Dominating Turkic dynastic tribe Sary (Ch. Se).
On another note:
The Ashina chose Tocharin women as dynastic mates, whereby, the pattern would indicate that the Ashina
males had to be either Q1a2 or Q1b. In other words, Ashina Y-DNA should be Q1a2 or Q1b, because it
states that the Turks followed that dynastic pattern.
The only exception to this (and may be not based upon the haplogroup(s) in question) would be the dynastic partners (brother-in-law material) for Ashina women, who it's stated were from the Barc tribe/clan?
If this issue is resolved, and Q1a2 is ruled out, theneverything points to Fred's theory as being correct. Even with these two issues, everything is pointing towards his theory as being correct. These two issues are really just loose ends.
I think that Fred's theory is right on.
[Note: With ancient history sources, it's not possible to scientifically triangulate the data. However, it is possible to cross-reference the data, but the expert validity of the sources is not always made clear...As in, are they quoting unreferenced credible sources, or not? In any event, the best way around that is to cross-reference the data from credible sources (but that is not always possible), and to triangulate scientific data, wherever possible, with
those sources.]
A Follow up Note on Dynastic Mating Patterns of the Xiongnu.
By the 3rd century BCE, the upper stratum of the Xiongnu was made up of five aristocratic houses:
Luandi/Luanti (house of Shanyu and the Tuqi), Huyan, Xubu, Qiulin, and Lan.
The Luandi (Luanti) was a clan and the ruling dynasty of the ancient Xiongnu that flourished
between 3rd century BCE to 4th century CE.
The Suibu was a maternal dynastic tribe of the ancient Eastern Huns (Ch. Xiongnu) that flourished between 3rd century BCE to 4th century CE.
In the Xiongnu federation, the dynastic clan was the Lunadi (Luant, Luanti) clan, which acted as a paternalistic dynastic tribe. The maternal dynastic tribes were the Huyan and Suibu. The Suibu tribe replaced the tribe Huyan, which was an earlier maternal dynastic tribe of the dynastic union with the paternal dynastic tribe Luanti.
The male members of the maternal dynastic line were not eligible for the Chanyu throne, only the male members of the Luanti line, whose father was a Luanti Chanyu, and mother was a Suibu Khatun (Queen) were eligible for the supreme
throne.
The Huyan tribe moved from the Right (Western) Wing, where the maternal dynastic tribe is traditionally assigned, to the Left (Eastern) Wing. (Huyan belonged to the dominating left wing, and Suibu belonged to the right wing.) Of the
noble tribes (other than Luanti), Huyan belonged to the dominating Left Wing, and Lan and Suibu belonged to the Right Wing.
http://en.wikipedia.org/wiki/Suibu_(tribe)
The Huns (Xiongnu/Huns)
----------------------------------------
The Hun tribes, or as the Chinese called them the Xiongnu, stemmed basically from the Siberian
branch of the Mongolian race.
http://www.silk-road.com/artl/xiongnu1.shtml
The Xiongu > The Huns
------------------------------------
Despite the fact that Mongolia is still one of the most isolated and one of the most sparsely populated countries in world, Y-DNA Q1b (haplogroup very rare in human populations for itself) has been detected by researchers from Department of Legal Medicine and Bioethics, Graduate School of Medicine, Nagoya University, Japan in all five regional populations (Ulaangom, Dalandzadgad, Ulaanbaatar,Undurkhaan,Choibalsan) in Mongolia and even in Japanese population (日本人 Nihonjin, Nipponjin).
Therefore this finding is of etreme importance due to the reason that near Mongolian southern border is location of cemetery Heigouliang, Xinjiang (Black Gouliang cemetery), east of Barkol basin, near the city of Hami. By typing results of DNA samples during the excavation of one of the tombs it was determined that of the 12 men there were: Q1a - 6, Q1b (M378) - 4, Q-2 (unable to determine subclade). Hosts of the tombs were representatives of Y-haplogroup Q1b-M378 exclusively (while Y-DNA Q1a represents sacrificial victims). They date from the time of early (Western) Han (2-1 century BC). In the very same location today Turkic Uygurs shows Q1b haplotypes very close if not the same as Ashkenazi Jewish Q1b(1a).
Source of attached table: http://www.fsigeneticssup.com/…/S1875-1768%2813%2…/fulltext…
Ashina was a branch of Xiongnu.
The Xiongnu established two dynasties. Both dynasties were organized as nomads. Among the united tribes there was
supposedly a tribe called Ashina (which was the name of the then leading clan of the steppe nomads or Turk (Turkut).
It's stated that the Ashina were related to the Northern branch of the Xiongnu.
(According to the New Book of Tang, the Ashina were related to the northern tribes of the Xiongnu, in particular they were of Tiele tribe by ancestral lineage.)
(See the part, below, on Hun Dynastic mating practices.)
It's further stated that the Northern branch of the Xiongnu, left the federation and become known as the Huns.
The Huns are Q1a2 and Q1b
------------------------------------------
This material is taken directly from this site, with limited paraphrasing:
Quotes:
Other Terms for Huns
---------------------------------
In the earlier pre-historic period the Huns were called Hu.
In the late pre-historic period the Huns were called Hun-yui; during the Zhou period (1045–256 BC) they were called Hyan-yun; starting from the Qin period (221-206 BC) they were called Hunnu.
The Tele Tribes
------------------------
The Tele (Tiele) were a branch of the Huns.
The Tele tribes emerged after the disintegration of the Xiongnu confederacy.
The Tele left the early Huns Horde, keeping patriarchal relations and a nomadic life.
The Tele were a collection of tribes of different Turkic ethnic origins.
The leaders of the Hun tribe had a greater level of mongoloidness than that of the members of the Tele (estimated from biological studies, as reaching 89%).
http://webcache.googleusercontent.com/search…
Tele Tribes and Uigurs
-----------------------------------
Besides that, the known ancient written sources indicate the origin of some Turkic-speaking tribes (belonging to the Tele group) can be traced directly from the Huns. Take the Uigurs...Tangshu directly tells that Uigur ancestors were Huns. In the earlier dynastic history, the ancestors of the Tele tribes, in particular the Uigurs, are derived from the Huns.
(Note: This fits with the Hun Dynastic model.See the Hun Dynastic Model.)
The nearest historical ancestors of modern Southern Altaians were mainly Tele tribes, and four centuries ago coached in the territory of Siberia, not only in its southern, mountain part in the Sayano-Altai mountains, but also in
the forest-steppes.
The so-called "real Huns" during the Ancient Turkic time have not disappeared at all. A number of tribes from the ethnic composition of the Huns have joined, probably partially, under their tribal names, partially under changed names, in the confederation of the Tele tribes, or in the Ancient Turkic Kaganates. Among the Tele tribes, one tribe even had the folk name of Hunnu (Huns) and it's known that it was called Hun.
Mongoloid Descent Noted
----------------------------------------
Turkic So, was stated to be the primogenitor tribe of Turk tribes.
The Turkic Tele Uigur clan, Toba, was also stated to be the founder dynastic clan. Toba descended from the tribe/country So (So-lu), which, as was stated, was the progenitor ancestor of the Turks.
Toba were Tungus Tanguts = future Mongol Uhuans (Wuhuan).
The Dulo was the Tele Tribes
--------------------------------------------
The Tokuz-Oguzes are called "nine tribes", which is a translation into the Chinese language of the term Tokuz-Oguz. The name Tokuz-Oguz means a confederation of the 9 Tele tribes.
In Middle Asia "Tele" was pronounced as "Dulo", and in that form it appears in the dynastic and historical records.
The Tele Kaganate was headed by Seyanto leaders. Uigurs also played a role.
The political influence of the (Tele) Kaganate headed by Seyanto extended to the territory of the modern Mongolia, Tuva, Altai, and the Hyagas state, located in the Minusinsk depression.
At the end of the 6th century the Western Turks controlled the lands north from the Gobi, and a majority of the Tele tribes fell under their rule.
Conflict between the Ashina/China and
-----------------------------------------------------------
Seyanto (of the Tele tribes)
-----------------------------------------
The tribal aristocracy of the eastern Turks, subordinated to China, elected as a leader Kupi, from the Kagan clan Ashina.
In 632 to 635 Teles crushed the Western Turkic Kagans. The Tokuz-Oguzes were led by Seyanto.
Kupi established his court to the north of the Mongolian Altai, where he proclaimed himself Turkic Kagan.
In 646 the Chinese army, together with the calvary of the eastern Turks trounced Seyanto. Seyanto was completely broken and dispersed.
http://s155239215.onlinehome.us/…/201Alt…/Potapov-TeleEn.htm
The Tele and the Ashina
-------------------------------------
There appear to be no written records that list the Ashina and Dulo as being listed as being one of the 12 or 15 tribes of the Tele Tribes.
Although there is this...
Quote
Bichurin 1.4 recites 12 Tele ancestral tribes in Ch. rendition see analysis by Gumilev:
The first two...
1, Lifuli (??????) (Barsil? "fu-li" is "bori"
= Tr. wolf)
2. Tulu (????) (Tunlo? Dulo? Dubo?)
However, there are also references that associate the Ashina with with Tele tribes.
Note that the Dulo and the Ashina usedthe same tagma symbol.
However, the Ashina were not actually listed as being one of the 12 or 15 tribes of the Tele tribe. Therefore one has to interpret the term "...they were of the Tiele tribe by ancestral lineage."
(See the part on Hun Dynastic mating
practices.)
The Hun's Dynastic Clan
--------------------------------------
The Hun's dynastic clan was Uiguro-Hunnic, or Tele-Hunnic. They were the Hun tribe's papas and Tele/Uigur tribe's mamas. The tribes of Huns and Tele/Uigurs were permanent mutual conjugal partners, a custom retained among the Turkic peoples up to the present time, and a major tool in a scientific research.
Hun tribe's Papas + Tele/Uigur tribe's Mamas
The Hun/Uigur conjugal union ascends to the time before their appearance in the Chinese annals, and the looks of the dynastic Huns and Uigurs was sufficiently blended after centuries, if not a millennium, of confined cross-breeding.
http://webcache.googleusercontent.com/search…
Notes:
Uyghur/Uigur--Originally known as the Dingling, an ancient Siberian people.
The Uigur's ancestors were Huns:
Uigur ancestors were Huns. In the earlier dynastic history, the ancestors of the Tele tribes (The Dulo), in particular the Uigurs, are derived from the Huns.
(Note: This fits with the Hun dynastic model.)
The Ashina were Huns
-----------------------------------
Among the united tribes, of the Xiongnu federation, there was a tribe called Ashina, which was the name of the leading clan of the steppe nomads (Turk).
From the dynastic history, it was already known that the ancestors of the ancient Turks, under the name Ashina, were a separate branch of the house of Hunnu (Huns).
The ethnogenetical connection of the ancient Turks with the Hunnu (Huns) was definitely stated by the source.
The ancestors of the ancient Turks, under a name Ashina, were a separate branch of the house of Hunnu (Huns)."
-- the Zhou (551–583) dynastic history
The Ashina were actually Huns, and Iranian
------------------------------------------------------------------
Barbarians
-----------------
According to the New Book of Tang, the Ashina were related to the northern tribes of the Xiongnu, in particular they were of the Tele tribe by ancestral lineage.
(See the part on Hun Dynastic mating practices.)
According to the Book of Zhou and History of Northern Dynasties, the Ashina were a branch of the Xiongnu, while according to the Book of Sui and Tongdian they were "a mixture of Hu" from Pingliang.
(Note: Hu means Huns.)
(See the part on Hun Dynastic mating
practices.)
There is no doubt that the Turks came from a mixing of the late Huns, who penetrated to the west after 265 AD (during a mass migration to the west of the Hunnic tribes from Central Asia), in the area of the small Pinlyan and Hesi states, with the local Iranian-lingual "barbarians."
Hun Tribes
The Ashina Turk type of the Huns descended from the Hun tribe Se/So.
Ashina was a "member tribe" of the So/Se/Sek/Saka tribes.
So/Se/Sek/Saka was the Parental tribe of the Ancient Turks.
The Saka were a Scythian tribe or group of tribes of Iranian origin.
Turkic So, was stated to be the primogenitor tribe of Turk tribes.
From there, after a defeat of Pinlyan by China in the 2nd half of the 5 century, the ancestors of the Turks went to the Gaochan mountains, and were subjugated by Jujans, and then settled by Jujans on the southern slopes of Mongolian Altai, where they were engaged in metalwork for the Jujan kagans.
Notes:
The migration of the Turks occurred under the Asina clan (originally Hiung-nu, so Asiatic Hun).
More accurately, it's stated, the Hunnic tribe Saka [Saka/Se] is given by Chinese annals of the later Wei Dynasty.
http://webcache.googleusercontent.com/search?
q=cache:3cbOa1xTU4wJ:s155239215.onlinehome.
us/turkic/23Avars/InMemoryOfAvarKhagans.htm+
the+Hun+tribes+th+so+clan&cd=16&hl=en&ct=
clnk&gl=us
This makes sense in light of other information on the Ashina: It's stated that the Ashina Turk type of the Huns descended from the Hun tribe Se/So. Some authors have implied that there is an Iranian link between the Se/So and the
Ashina.
It's also stated that theTurks descended from the tribe So that lived in the northern Altai.
Hun's tribes--Dominating Turkic dynastic tribe Sary (Se).
Se > Chinese term.
Se = Sakas
It's stated that the Se-yanto or Syr-Tardush were an ancient Tele people.
Seyanto (Sir + Yanto)--The Syr-Tardush
In 631, the Seyanto state successfully controlled all Turk's leaders except for Ordos Turks under Chinese protection.
It's also stated that Seyanto and Bugu had identical tamgas. (Note so did the Ashina and the Dulo...? However, also note that the Tele was known as the Dulo in central Asia.)
Notes:
Although the Tele was also known as the Dulo, different sources also make distinctions:
The On Okh (Onoq, Ten Arrows) or Western Turks--Subdivision: The greater Tardush (Tulu, Dulo) <----
---Branch: Turgesh
The Turgesh were a Turkic tribal confederation who emerged from the ruins of the Western
Turkic Kaganate.
This indicates that the greater Tardush was the Tulu, Dulo clan.
http://en.wikipedia.org/wiki/Turgesh
The Turgesh were a group of Duolu tribes .
The Foundation of the Turgesh Kaganate was Precipitated by Anti- Ashina Turgesh rebellion. Ashina--The Counter-Movement of Turgeshes, ended with a capture of six hundred and ninety-nine in Suyab .
http://translate.google.com/translate…
Notes:
The Dulo and the Ashina were also referred to as being cousins, in other sources, who became opposed to each other, as the result of ethnic tensions.
The Khazar kingdom was ruled by the Ashina dynasty of Western Turks (Turkuts), who took over the territory.
The On Okh (Onoq, Ten Arrows) or Western Turks--Subdivision: The greater Tardush (Tulu, Dulo) <----
---Branch: Turgesh
http://www.jstor.org/discover/10.2307/41035790…
Comments
-----------------
The Xiongu > The Huns (Q1a2 and Q1b) > Ashina
(See the part on Hun Dynastic mating practices.)
It appears that there was both a Q1a2 and a Q1b genetic core in the Huns, and based upon the Hun dynastic practices of mating, it appears that the Pappas would continue to be Q1a2 and Q1b, and that the Mammas came from the Tele tribes.
Being related to the Tele tribe tells one nothing specific, about haplogroups, but the presence of Q is indicated through the Ashina being related to the Northern branch of the Xiongnu, who became the Huns.
The Ashina were a separate branch of the house of Hunnu (Huns).
The Ashina were related to the Northern Branch of the Xiongnu/Huns who became known as the Northern Huns.
According to the Book of Zhou and History of Northern Dynasties, the Ashina were a branch of the Xiongnu, while according to the Book of Sui and Tongdian they were "a mixture of Hu" (Huns) from Pingliang.
According to the New Book of Tang, the Ashina were related to the northern tribes of the Xiongnu, in particular they were of the Tele tribe by ancestral lineage.
The Tele also left the Huns, keeping patriarchal relations.
The custom (Hun dynastic mating practices) was retained among the Turkic peoples up to the present time. Therefore, this implies that Hun Y-DNA (Q1a2 and Q1b) was passed down, throughthe Ashina males, and that the Turkic peoples
retained this custom.
Research has indicated that the Northern Huns genetic core was Q1a2 and Q1b.
Huns (Y: Q1a2 and Q1b) > Tele (MtDNA)
According to the Book of Sui and Tongdian the Ashina were a mixture of Hu (Huns) from Pingliang.
There is no doubt that the Turks came from a mixing of the late Huns, in the area of the small Pinlyan and Hesi states, with the local Iranian-lingual "barbarians."
Certain authors have traced Se/So/Saka to Iranian sources. What appeared to be contradictory material makes sense in light of the Hun dynastic mating model, because it connects all of this material together.
The Ashina were a seperate branch of the House of Huns. The ancestors (Ashina) of the Turks were a branch of the Huns.
The Ashina were the result of the intermixing of the Huns with Iranian Barbarians.
The view widely spread in the world of Oriental studies is that that Huns were ancestors of the ancient Turks as was repeatedly certified by the ancient Chinese written sources.
From the dynastic history it's known that the ancestors of the ancient Turks, under a name Ashina, were a separate branch of the house of Hunnu (Huns). The ethnogenetical connection of the ancient Turks with the Huns was definitely
stated by the source. But there is other historical information that allows one to detail and refine this connection.
There is no doubt that they came from a mixing of the late Huns, in the area of the small Pinlyan and Hesi states, with the local Iranian-lingual barbarians. From there, after a defeat of Pinlyan by China, the ancestors of the Turks coached
away to the Gaochan mountains, northwest from Turfan, and were subjugated by Jujans, and then settled by Jujans on the southern slopes of Mongolian Altai, where they engaged in metalwork for the Jujan kagans.
http://webcache.googleusercontent.com/search…
(This particular example of intermixing, applied to the creation of the Ashina, would explain why one can trace the Ashina origins in two directions. In light of this material, it's not a contradiction, as it is missing pieces of a puzzlethat puts everything into perspective by making it fit together.)
If the Tele was the Dulo, as was claimed, then, based upon the Hun dynastic model, it wouldindicate that the Northern Huns would have been Q1a2 and Q1b...Hence, the Ashina would have been Q1a2 or Q1b...Also, Atiila the Hun
was associated with Dulo (Tele Tribes), and according to research, Atiila the Hun was believed to be either Q1a2 or Q1b. That would also fit with the Hun dynastic model.
Ashina Dynastic Partners
---------------------------------------
The female dynastic partner of the Ashina Turks, with a title to the country land and people, is a pin. Yuezhi tribe, which is a Chinese codeword for the Tokhars.
http://s155239215.onlinehome.us/tu…/29Huns/CsornaiHunsEn.htm
Tahia (Tokhars)
The Yuezhi may have been a Europoid people, as indicated by the portraits of their kings on the coins they struck following their exodus to Transoxiana (2nd-1st century BCE)
Comments
-----------------
It further states that the Turks continued this dynastic practice. Therefore, it appears that Pappa Hun Y-DNA would have to continue to be either Q1a2 or Q1b. It appears that the Ashina (Hun) Pappas dynastic practices eventually involved mating with Tokhar women instead of women from the Tele tribe. This would indicate that the male descendants of Ashina males would continue to be either Q1a2 or Q1b.
Based upon these patterns, it appears thatthe only other Y-DNA haplogroup, that could be introduced into the Ashina line, would bethrough the dynastic partners of the Ashinawomen, who were from the Barc tribe, and
that is only if they weren't also Q1a2 or Q1b.
(Note the Barc tribe/clan was stated to havebeen brother-in-law material for the Ashina women.)
http://www.jstor.org/discover/10.2307/41035790…
Huns (Y-DNA: Q1a2 and Q1b) > Tele (MtDNA)
Hun/Ashina (Y-DNA: Q1a2 and Q1b) > Tokhars (MtDNA)
The following factor needs to be factored into this:
Ashina Women > Barc Tribe Males (Y: ?)
Notes for it are:
The non-qaganal clan of the Barc/Warâz/Bolcân, or VarazAVarac/Barc, or Varaz/Warac/Barc, who were another western turk clan.
nekagansky clan Bartsch / Varaz / Bolchan ( Barc / Waraz / Bolcan ) - corresponding dynasty Ihshad / Running / Iillig (Ihsed / Beg / Yillig)
Comments
-----------------
The Huns were Q1a2 and Q1b.
Atilla the Hun was from the Dulo (Tele) and he was believed to be Q1a2 or Q1b.
The Huns dynastic model involved mating practices where the Huns were the Pappas (Y-DNA Q1a2 or Q1b)
and the Mammas were from the Tele/Uigurs (mtDNA).
The Ashina were the result of Hun Y-DNA + Iranian Barbarian mtDNA. The Ashina were also associated
with the Tele.
Other sources connect the Ashina with So/Se, which is connected to Iranian sources, and this starts
making sense, in light of the above information.
Tele Tribes
1 Uange (Uygurs)
2 Seyanto (Sir + Yanto)
3 Kibi (Kibir)
4 Dubo (Tubalar)(Dabo)(Tele)
5 Guligan (Kurykan)(Yakut)
6 Dolange (Telengits)
7 Bugu (Pugu)(Uygurs)
8 Bayegu (Baiyrku)(Uygurs)
9 Tunlo (Tongra)(Uygurs)
10 Hun
11 Sygye (Uygurs)
12 Husye
13 Higye
14 Adye(Eduz)
15 Baysi (Barsil)
http://s155239215.onlinehome.us/…/71_…/hun_dateline_1_En.htm
It's stated that theTurks descended from the tribe So that lived in the northern Altai.
In the Hun's tribes, it's been stated that the dominating Turkic dynastic tribe was Sary
(Se).
Se > Chinese term.
There are also clans in tribe Se.
--A clan of tribe Se (Sir/Sary).
In 300 AD, Sirs and Turks lived at Ordos.
http://s155239215.onlinehome.us/…/71_…/hun_dateline_1_En.htm
It's stated that the Se-yanto or Syr-Tardush were an ancient Tele people.
Seyanto (Sir + Yanto)--The Syr-Tardush
It's stated that Alat-tou belonged to a clan oftribe Se (Sir/Sary), who at some time wiped out Yanto leadership, and took over leadership of the Sary/Yanto confederation.
Dominating Turkic dynastic tribe Sary (Ch. Se).
On another note:
The Ashina chose Tocharin women as dynastic mates, whereby, the pattern would indicate that the Ashina
males had to be either Q1a2 or Q1b. In other words, Ashina Y-DNA should be Q1a2 or Q1b, because it
states that the Turks followed that dynastic pattern.
The only exception to this (and may be not based upon the haplogroup(s) in question) would be the dynastic partners (brother-in-law material) for Ashina women, who it's stated were from the Barc tribe/clan?
If this issue is resolved, and Q1a2 is ruled out, theneverything points to Fred's theory as being correct. Even with these two issues, everything is pointing towards his theory as being correct. These two issues are really just loose ends.
I think that Fred's theory is right on.
[Note: With ancient history sources, it's not possible to scientifically triangulate the data. However, it is possible to cross-reference the data, but the expert validity of the sources is not always made clear...As in, are they quoting unreferenced credible sources, or not? In any event, the best way around that is to cross-reference the data from credible sources (but that is not always possible), and to triangulate scientific data, wherever possible, with
those sources.]
A Follow up Note on Dynastic Mating Patterns of the Xiongnu.
By the 3rd century BCE, the upper stratum of the Xiongnu was made up of five aristocratic houses:
Luandi/Luanti (house of Shanyu and the Tuqi), Huyan, Xubu, Qiulin, and Lan.
The Luandi (Luanti) was a clan and the ruling dynasty of the ancient Xiongnu that flourished
between 3rd century BCE to 4th century CE.
The Suibu was a maternal dynastic tribe of the ancient Eastern Huns (Ch. Xiongnu) that flourished between 3rd century BCE to 4th century CE.
In the Xiongnu federation, the dynastic clan was the Lunadi (Luant, Luanti) clan, which acted as a paternalistic dynastic tribe. The maternal dynastic tribes were the Huyan and Suibu. The Suibu tribe replaced the tribe Huyan, which was an earlier maternal dynastic tribe of the dynastic union with the paternal dynastic tribe Luanti.
The male members of the maternal dynastic line were not eligible for the Chanyu throne, only the male members of the Luanti line, whose father was a Luanti Chanyu, and mother was a Suibu Khatun (Queen) were eligible for the supreme
throne.
The Huyan tribe moved from the Right (Western) Wing, where the maternal dynastic tribe is traditionally assigned, to the Left (Eastern) Wing. (Huyan belonged to the dominating left wing, and Suibu belonged to the right wing.) Of the
noble tribes (other than Luanti), Huyan belonged to the dominating Left Wing, and Lan and Suibu belonged to the Right Wing.
http://en.wikipedia.org/wiki/Suibu_(tribe)
Q1b Genetics
The update of the phylogenetic structure of Q1b haplogroup based on full Y-chromosome sequencing
Vladimir Gurianov Roman Sychyev Vladimir Tagankin Vadim Urasin 1 Independent Researcher, Russia,
Abstract The new data of full Y-chromosome sequencing allowed the update of the Q1b (Q-L275) haplogroup struc-ture, as well as in identifying new subclades: Q-Y2990 (downstream Q-Y2250), Q-Y2225 (downstream Q-Y2220) and Q-Y3030 (downstream Q-Y2200). It created the background for continuation of further researches of the in-ner structure of the pointed subclades and on comparing of their existing ethno-population composition with the migration of the Indo-European tribes.
https://www.academia.edu/7117246/The_update_of_the_phylogenetic_structure_of_Q1b_haplogroup_based_on_full_Y-chromosome_sequencing
Vladimir Gurianov Roman Sychyev Vladimir Tagankin Vadim Urasin 1 Independent Researcher, Russia,
Abstract The new data of full Y-chromosome sequencing allowed the update of the Q1b (Q-L275) haplogroup struc-ture, as well as in identifying new subclades: Q-Y2990 (downstream Q-Y2250), Q-Y2225 (downstream Q-Y2220) and Q-Y3030 (downstream Q-Y2200). It created the background for continuation of further researches of the in-ner structure of the pointed subclades and on comparing of their existing ethno-population composition with the migration of the Indo-European tribes.
https://www.academia.edu/7117246/The_update_of_the_phylogenetic_structure_of_Q1b_haplogroup_based_on_full_Y-chromosome_sequencing
ASHINA TRIBE WAS XIONGNU/HUNNU/HUNS
"The ancestors of the ancient Turks, under a name Ashina, were a
separate branch of the House of Hunnu (Huns)."
-- the Zhou (551–583) Dynastic history
Chinese archaeologists have published several reviews regarding the results of excavations in Xinjiang. Particularly interesting are in the cemetery Heigouliang, Xinjiang (Black Gouliang cemetery), east of BARKOL basin, near the city of Hami. By typing results of DNA samples during the excavation of one of the tombs it was determined that of the 12 men there were: Q1a - 6, Q1b (M378) - 4, Q-2 (unable to determine subclade). Hosts of the tombs were representatives of Y-haplogroup Q1b-M378 exclusively (while Y-DNA Q1a represents sacrificial victims). They date from the time of early (Western) Han (2-1 century BC). Summarizing the data from available evidences, it is concluded that the tomb belongs to the representatives of the Xiongnu/Hunnu nobility/conquerors.[118][119][120]
http://en.wikipedia.org/wiki/Xiongnu#Genetics
"The ancestors of the ancient Turks, under a name Ashina, were a
separate branch of the House of Hunnu (Huns)."
-- the Zhou (551–583) Dynastic history
Chinese archaeologists have published several reviews regarding the results of excavations in Xinjiang. Particularly interesting are in the cemetery Heigouliang, Xinjiang (Black Gouliang cemetery), east of BARKOL basin, near the city of Hami. By typing results of DNA samples during the excavation of one of the tombs it was determined that of the 12 men there were: Q1a - 6, Q1b (M378) - 4, Q-2 (unable to determine subclade). Hosts of the tombs were representatives of Y-haplogroup Q1b-M378 exclusively (while Y-DNA Q1a represents sacrificial victims). They date from the time of early (Western) Han (2-1 century BC). Summarizing the data from available evidences, it is concluded that the tomb belongs to the representatives of the Xiongnu/Hunnu nobility/conquerors.[118][119][120]
http://en.wikipedia.org/wiki/Xiongnu#Genetics
While Q1a is more Mongolian, Siberian and Native American, Q1b (L275) appears to have originated in Central Asia and migrated early to South Asia and the Middle East. The highest frequency of Q1b in Europe is found among Ashkenazi Jews (5%) and Sephardic Jews (2%), suggesting that Q1b was present in the Levant before the Jewish disapora 2,000 years ago. Q1b is also found in Lebanon (2%), and in isolated places settled by the Phoenicians in southern Europe (Crete, Sicily, south-west Iberia). This means that Q1b must have been present in the Levant at latest around 1200 BCE, a very long time before the Hunnic migrations. One hypothesis is that Q1b reached the Middle East alongside haplogroupR1a-Z93 with the Indo-Iranian migrations from Central Asia during the Late Bronze Age.
Q1b may almost certainly not one of the original lineages of Proto-Indo-European speakers of the Pontic-Caspian Steppe since it is almost completely absent from Balto-Slavic and Germanic countries. Nevertheless, it is reasonable to assume that Q1b was indigenous to Central Asia and was absorbed by the Indo-Iranian branch of the Indo-Europeans there during the Bronze Age Andronovo culture, then spread with the Indo-Aryans to India, Iran and the Near East. Q1b probably settled in the Levant at the same time as R1a-Z93, as both lineages are found among the Jews and the Lebanese and in places historically colonised by the Phoenicians. Autosomal analyses have confirmed that all Levantine people (Jews, Lebanese, Palestinians, Syrians) possess about 0.5% of Northeast Asian (Mongoloid) admixture. Since these populations lack Mongoloid mtDNA, the presence of Northeast Asian admixture can only be explained by the 2% of Q1b among Levantine men, the only paternal lineage of Mongoloid origin in the region.
Q1b may almost certainly not one of the original lineages of Proto-Indo-European speakers of the Pontic-Caspian Steppe since it is almost completely absent from Balto-Slavic and Germanic countries. Nevertheless, it is reasonable to assume that Q1b was indigenous to Central Asia and was absorbed by the Indo-Iranian branch of the Indo-Europeans there during the Bronze Age Andronovo culture, then spread with the Indo-Aryans to India, Iran and the Near East. Q1b probably settled in the Levant at the same time as R1a-Z93, as both lineages are found among the Jews and the Lebanese and in places historically colonised by the Phoenicians. Autosomal analyses have confirmed that all Levantine people (Jews, Lebanese, Palestinians, Syrians) possess about 0.5% of Northeast Asian (Mongoloid) admixture. Since these populations lack Mongoloid mtDNA, the presence of Northeast Asian admixture can only be explained by the 2% of Q1b among Levantine men, the only paternal lineage of Mongoloid origin in the region.
JEWISH SOURCES ABOUT HUN ORIGIN OF KHAZARS
JEWISH VIRTUAL LIBRARY:
The Origin of the Khazars
The Khazars, of Turkic stock, originally nomadic, reached the Volga-Caucasus region from farther east at some time not easily determinable.
They may have belonged to the empire of the Huns (fifth century C.E.) as the Akatzirs, mentioned by Priscus.
This name is said to be equivalent to Aq-Khazar, i.e., White Khazars, as opposed to the Qara-Khazar or Black Khazars mentioned by al-Iṣṭakhrī (see below).
The Khazars probably belonged to the West Turkish Empire (from 552 C.E.), and they may have marched with Sinjibū (Istämi), the first khāqān of the West Turks, against the Sassanid (Persian) fortress of Ṣul or Darband
In the time of Procopius (sixth century) the region immediately north of the Caucasus was held by the Sabirs, who are referred to by Jordanes as one of the two great branches of the Huns (Getica, ed. Mommsen, 63). Masʿūdī (tenth century C.E.) says that the Khazars are called in Turkish, Sabīr (Tanbīh, ed. Cairo, 1938, 72).
In 627 (Theophanes, Chronographia, ed. De Boor, 1 (1883), 315) "the Turks from the East whom they call Khazars" under their chief, Ziebel, passed the Caspian Gates (Darband) and joined Heraclius at the siege of Tiflis. In view of what is known of a dual kingship among the Khazars (see below), it would be natural to assume that Ziebel, described by Theophanes as "second in rank to the khāqān," was the subordinate Khazar king or beg. However, there are grounds for thinking that Ziebel stands for yabgu, a Turkish title – in the parallel Armenian account (Moses of Kalankatuk, trans. Dowsett, 87) he is called Jebu Khāqān – and that he is T'ung-ye-hu, Ye-hu Khagan of the Chinese sources, i.e., T'ung Yabgu, Yabgu Khāqān, the paramount ruler of the West Turks, who is represented as second in rank to "the King of the North, the lord of the whole world," i.e., the supreme khāqān of the Turks. In the narratives of Theophanes and Moses of Kalankatuk respectively, the Khazars are also called Turks and Huns. From 681 C.E., we hear much in the latter author of the Huns of Varach ʿ an (Warathān), north of Darband, who evidently formed part of a Khazar confederation or empire. Their prince Alp Ilutver was often in attendance on the Khazar khāqān and was converted to Christianity by an Albanian bishop.
http://www.jewishvirtuallibrary.org/...m/khazars.html
HUNS IN MESOPOTAMIA, ANATOLIA AND LEVANT IN 395/396-398 AC
In my own Y-DNA results at FTDNA I have noted a very few, but present non Jewish matches in Iran, Iraq, Turkey/Armenia.
I think that Encyclopedia Iranica provides interesting insight when it comes to the description of the "European Huns" activity in Near East:
.....
A direct confrontation between the Huns and the Persian empire first occurred twenty years after the beginning of the great migration. In the summer of 395, hordes of Huns crossed the Don near its estuary, turned southeast, and made their way through the Caucasus into Persia and the Roman provinces. While the plundering of the Roman areas is variously attested (for sources, see Maenchen-Helfen, pp. 38-42), only Priscus (frag. 11) and the Liber Calipharum (Chronicon miscellaneum 3.4; tr., pp. 106-7) report the invasion of the Persian empire. Under the leadership of Basikh and Koursikh, a detachment of Huns rode down the valleys of the Euphrates and Tigris to Ctesiphon. Upon the news that a Persian army was marching towards them, the Huns turned back; but they were eventually caught. The Persians then managed to kill some of the Huns, to take almost their entire booty from them, and allegedly to free 18,000 prisoners. The rest of the Huns troops made their way back into the steppe over the Darband pass. This inroad into Persia was remembered by the Romans and the Huns more than fifty years later: when Priscus was staying at Attila’s court in 449 C.E., he heard from the Western Roman envoy Romulus that the king was planning a campaign against Persia, which was to be carried out on the route previously taken by Basikh and Koursikh (Priscus, frag. 11). Attila’s death in 453 C.E. saved the Sasanians from an armed encounter with the Huns while they were at the height of their military power.
.....
A tribe which soon after 500 C.E. invaded northern Iran and was simply called “Huns” (Ounnoi) by Procopius (Bella 1.8) might be identified with the Sabires themselves, who from then on participated in the Persian-Byzantine wars for several decades, siding alternately with the Persians and the Eastern Romans. In the conflicts of the mid-6th century, both warring parties were supported by Sabire detachments. After having fought on the Persian side in 573 C.E., the Sabires subjected themselves a year later to the Eastern Roman emperor and were settled in the Byzantine part of Armenia (Moravcsik, I, p. 68).
http://www.iranicaonline.org/articles/huns
The Cambridge History of Early Inner Asia expands previous information with the fact that Hun Army in 395-396 (THE GREAT HUN RAID) under leadership of BASIKH and KOURSIKH (arkhontes-high ranking military commanders) also Armenia, Syria, Palestine (!) and Northern Mesopotamia.
http://librarum.org/book/38133/192
JEWISH VIRTUAL LIBRARY:
The Origin of the Khazars
The Khazars, of Turkic stock, originally nomadic, reached the Volga-Caucasus region from farther east at some time not easily determinable.
They may have belonged to the empire of the Huns (fifth century C.E.) as the Akatzirs, mentioned by Priscus.
This name is said to be equivalent to Aq-Khazar, i.e., White Khazars, as opposed to the Qara-Khazar or Black Khazars mentioned by al-Iṣṭakhrī (see below).
The Khazars probably belonged to the West Turkish Empire (from 552 C.E.), and they may have marched with Sinjibū (Istämi), the first khāqān of the West Turks, against the Sassanid (Persian) fortress of Ṣul or Darband
In the time of Procopius (sixth century) the region immediately north of the Caucasus was held by the Sabirs, who are referred to by Jordanes as one of the two great branches of the Huns (Getica, ed. Mommsen, 63). Masʿūdī (tenth century C.E.) says that the Khazars are called in Turkish, Sabīr (Tanbīh, ed. Cairo, 1938, 72).
In 627 (Theophanes, Chronographia, ed. De Boor, 1 (1883), 315) "the Turks from the East whom they call Khazars" under their chief, Ziebel, passed the Caspian Gates (Darband) and joined Heraclius at the siege of Tiflis. In view of what is known of a dual kingship among the Khazars (see below), it would be natural to assume that Ziebel, described by Theophanes as "second in rank to the khāqān," was the subordinate Khazar king or beg. However, there are grounds for thinking that Ziebel stands for yabgu, a Turkish title – in the parallel Armenian account (Moses of Kalankatuk, trans. Dowsett, 87) he is called Jebu Khāqān – and that he is T'ung-ye-hu, Ye-hu Khagan of the Chinese sources, i.e., T'ung Yabgu, Yabgu Khāqān, the paramount ruler of the West Turks, who is represented as second in rank to "the King of the North, the lord of the whole world," i.e., the supreme khāqān of the Turks. In the narratives of Theophanes and Moses of Kalankatuk respectively, the Khazars are also called Turks and Huns. From 681 C.E., we hear much in the latter author of the Huns of Varach ʿ an (Warathān), north of Darband, who evidently formed part of a Khazar confederation or empire. Their prince Alp Ilutver was often in attendance on the Khazar khāqān and was converted to Christianity by an Albanian bishop.
http://www.jewishvirtuallibrary.org/...m/khazars.html
HUNS IN MESOPOTAMIA, ANATOLIA AND LEVANT IN 395/396-398 AC
In my own Y-DNA results at FTDNA I have noted a very few, but present non Jewish matches in Iran, Iraq, Turkey/Armenia.
I think that Encyclopedia Iranica provides interesting insight when it comes to the description of the "European Huns" activity in Near East:
.....
A direct confrontation between the Huns and the Persian empire first occurred twenty years after the beginning of the great migration. In the summer of 395, hordes of Huns crossed the Don near its estuary, turned southeast, and made their way through the Caucasus into Persia and the Roman provinces. While the plundering of the Roman areas is variously attested (for sources, see Maenchen-Helfen, pp. 38-42), only Priscus (frag. 11) and the Liber Calipharum (Chronicon miscellaneum 3.4; tr., pp. 106-7) report the invasion of the Persian empire. Under the leadership of Basikh and Koursikh, a detachment of Huns rode down the valleys of the Euphrates and Tigris to Ctesiphon. Upon the news that a Persian army was marching towards them, the Huns turned back; but they were eventually caught. The Persians then managed to kill some of the Huns, to take almost their entire booty from them, and allegedly to free 18,000 prisoners. The rest of the Huns troops made their way back into the steppe over the Darband pass. This inroad into Persia was remembered by the Romans and the Huns more than fifty years later: when Priscus was staying at Attila’s court in 449 C.E., he heard from the Western Roman envoy Romulus that the king was planning a campaign against Persia, which was to be carried out on the route previously taken by Basikh and Koursikh (Priscus, frag. 11). Attila’s death in 453 C.E. saved the Sasanians from an armed encounter with the Huns while they were at the height of their military power.
.....
A tribe which soon after 500 C.E. invaded northern Iran and was simply called “Huns” (Ounnoi) by Procopius (Bella 1.8) might be identified with the Sabires themselves, who from then on participated in the Persian-Byzantine wars for several decades, siding alternately with the Persians and the Eastern Romans. In the conflicts of the mid-6th century, both warring parties were supported by Sabire detachments. After having fought on the Persian side in 573 C.E., the Sabires subjected themselves a year later to the Eastern Roman emperor and were settled in the Byzantine part of Armenia (Moravcsik, I, p. 68).
http://www.iranicaonline.org/articles/huns
The Cambridge History of Early Inner Asia expands previous information with the fact that Hun Army in 395-396 (THE GREAT HUN RAID) under leadership of BASIKH and KOURSIKH (arkhontes-high ranking military commanders) also Armenia, Syria, Palestine (!) and Northern Mesopotamia.
http://librarum.org/book/38133/192
Qabalistic Suncode of Genetic Programming, Iona Miller, 1982
The codones (four
genetic substances) should be read from the inside-out. The four
color-coded substances (G, A, U, C), combine first in 16 ways (4 x 4),
then in 64 ways (4 x 16). The magic number 64 immediately reminds
us of the 64 Hexagrams of the I Ching, the Chinese synergetic book of life.
In this painting, the Sun-Code occupies the place of Tiphareth, surrounded
by its satellite Spheres of the Tree of Life. The surrounding DNA
chain in the shape of the World Egg, is a variation of the alchemical tail-eating
serpent Ourobouros. Its head is formed by the Hebrew Yod,
a symbol of life and sperm, the unbroken circle of life.
HAPLOTYPE R1b Descent

"R1b is the haplogroup of the Pharoahs and their children, the Solomanic Kings of the Jews."--NDV
Haplogroup R1b is by far the most common haplogroup in western Europe (see map), yet everyone below has a rare haplotype. This rarity is a reflection of the fact that while haplogroup subclade R1b1b2 is the most common haplogroup in western Europe, the ancestral subclade, R1b1*, is quite rare.
HAPLOTYPE Y-DNA haplogroup R is believed to have arisen approximately 27,000 years ago in Asia. The two currently defined sublcades are R1 and R2. Haplogroup R1 is estimated to have arisen during the height of the Last Glacial Maximum (LGM), about 18,500 years ago, most likely in southwestern Asia. The two most common descendant clades of haplogroup R1 are R1a and R1b. R1a is believed to have arisen on the Eurasian Steppe, and today is most frequently observed in eastern Europe and in western and central Asia. R1b is believed to have arisen in southwest Asia and today is most frequently observed in Europe and especially in western Europe, which it entered after the LGM largely in the form of [[R1b1b2]]. The Atlantic Modal Haplotype, or AMH, is the most common STR haplotype in haplogroup [[R1b1b2a]]. R2 is most often observed in Asia, especially on the Indian sub-continent and in central Asia.
INFORMATION RESULTS FOR MARKERS IN STUDY GROUP :Y-SNP information for M269....SNPID-rs9786153 M269 marks the most common haplogroup found in Europe. Its frequency exceeds 80% in parts of western Europe and it occurs at lower frequencies in central and eastern Europe. It marks an ancient migration into Europe prior to the LGM and is believed to have survived in refugia in Iberia, the Balkans and possibly elsewhere before expanding northwards again. It apparently comprises two major branches defined by DYS1/p49a,f haplotypes 15 and 35 but this branching cannot yet be included in the tree because the underlying mutation is not verified as a unique event polymorphism. R1b1b2 ..... SNP M269
R1a - Originally thought to have originated in the Caucasus region around the Black Sea, new research is indicating that this type may have originated in the region around Khazakstan, possibly even in India or Pakistan. R1a spread into Central Asia and migrated across the Russian Steppes into Eastern Europe where it reaches high levels in Hungary, Poland, the Ukraine and the Slavic regions (the peoples genetically closest to Norwegians). R1a is characterized by the mutations SRY10831.2- (negative as opposed to positive) and M17+. M17 is what most academic studies have tested for to determine R1a - it actually categorizes R1a1, which seems to encompass all R1a (I have yet to see a single "R1a" that was SRY10831.2- and also M17-. Anyone SRY10831.2- seems to be universally M17+, in other words all R1a are also R1a1). For this reason you will many times see R1a and R1a1 used interchangably within the literature.
One particular group of Y-STR values within R1a shows matches in Central Asia, around the Siberian Altai and Uyghur province of Western China. The recent find of Caucasian mummies in the Takla Makan deserts of the Uyghur province prove that a race of red and blond haired people with Scandinavian features, over 6' tall, once lived in this region. R1a is found at very high percentages in Western Norway, where it reaches frequencies between 23% to 30%. Some researchers believe the Icelandic Sagas, which describe a migration of a population from Asia beyond the Ural mountains, to Norway, may actually be based in fact. Thor Heyerdahl, of Kon Tiki fame, spent the remaining years of his life attempting to prove this theory -- and DNA evidence is seeming to prove him right. The Swedes have long believed this legend, and the emergence of a specific type of Scandinavian R1a with a Central Asian motif seems to support this account.
The MacDonalds have determined that their progenitor, Somerled, belonged to haplogroup R1a (of the same Central Asian motif) and the Douglas's progenitor, Willem de Douglas, recently turned up R1a according to the Clan Douglas DNA project. It seems this holds true for most of the pseudo-aristocracy of Scandinavia. R1a is found at levels of less than 1% in most regions of Ireland, and at levels of 3-5% in England, and only slightly higher in Scotland. The highest concentrations of this haplogroup are seen in areas of Britain colonized by the Norse Vikings. One of the leading DNA experts has called R1a the only sure proof of Norse Viking origins when seen in men of deep British ancestry.
R1b - The most common haplogroup in Western Europe and Britain. This type is believed to have gestated in Spain during the Ice Age, migrating back into Europe with the retreat of the glaciers. People of Celtic ancestry show very close matches with the Basque people of Spain (more recent theories have divided R1b into "Iberian and Non-Iberian" and seem to indicate that some R1b spent the glacial period somewhere outside of Spain). Most people of European or British ancestry will fall into this haplogroup.
R1b is characterized by the P25 mutation (P25+). However, almost all R1b types fall within one of the sub-categories of R1b, typically M269+ called R1b3 (YCC 2003 Chart) or R1b1c (YCC 2005 Chart). This is a rapidly changing group and new SNP's are being discovered regularly so nothing here can be taken as static since it is very much fluid.
It is expected that most of the members of the Grant DNA Project will be of this haplogroup. In fact, the one line we have identified in our project as being the Chiefly line is R1b, which would be expected if the line were related to Andrew Stewart. The Royal line of the Stewarts in Scotland were descended from a family of Breton (in Western France) nobles, so it follows that if Andrew Stewart were of the Royal Line and married into the family, we should expect that the Chiefly line would be R1b. We have also identified a few kits in the project whose owners are directly related to the Chiefly line. [interpretations under review by the Court]
However, most of those that fall into the R1b haplogroup will be unrelated to either the current line of the Chiefs or the original Norse (Haplogroup R1a) line. These DNA signatures represent the various tacksmen, farmers/crofters, peasant classes, etc, that settled in Strathspey or Glenmoriston and assumed the name of the Chief on whose lands they were living. This is the reason that so many project members show very close matches with other individuals, of other surnames, even at the 25 or 37 marker levels in Family Tree DNA's "Y-Search" database.
Haplogroup R1b is by far the most common haplogroup in western Europe (see map), yet everyone below has a rare haplotype. This rarity is a reflection of the fact that while haplogroup subclade R1b1b2 is the most common haplogroup in western Europe, the ancestral subclade, R1b1*, is quite rare.
HAPLOTYPE Y-DNA haplogroup R is believed to have arisen approximately 27,000 years ago in Asia. The two currently defined sublcades are R1 and R2. Haplogroup R1 is estimated to have arisen during the height of the Last Glacial Maximum (LGM), about 18,500 years ago, most likely in southwestern Asia. The two most common descendant clades of haplogroup R1 are R1a and R1b. R1a is believed to have arisen on the Eurasian Steppe, and today is most frequently observed in eastern Europe and in western and central Asia. R1b is believed to have arisen in southwest Asia and today is most frequently observed in Europe and especially in western Europe, which it entered after the LGM largely in the form of [[R1b1b2]]. The Atlantic Modal Haplotype, or AMH, is the most common STR haplotype in haplogroup [[R1b1b2a]]. R2 is most often observed in Asia, especially on the Indian sub-continent and in central Asia.
INFORMATION RESULTS FOR MARKERS IN STUDY GROUP :Y-SNP information for M269....SNPID-rs9786153 M269 marks the most common haplogroup found in Europe. Its frequency exceeds 80% in parts of western Europe and it occurs at lower frequencies in central and eastern Europe. It marks an ancient migration into Europe prior to the LGM and is believed to have survived in refugia in Iberia, the Balkans and possibly elsewhere before expanding northwards again. It apparently comprises two major branches defined by DYS1/p49a,f haplotypes 15 and 35 but this branching cannot yet be included in the tree because the underlying mutation is not verified as a unique event polymorphism. R1b1b2 ..... SNP M269
R1a - Originally thought to have originated in the Caucasus region around the Black Sea, new research is indicating that this type may have originated in the region around Khazakstan, possibly even in India or Pakistan. R1a spread into Central Asia and migrated across the Russian Steppes into Eastern Europe where it reaches high levels in Hungary, Poland, the Ukraine and the Slavic regions (the peoples genetically closest to Norwegians). R1a is characterized by the mutations SRY10831.2- (negative as opposed to positive) and M17+. M17 is what most academic studies have tested for to determine R1a - it actually categorizes R1a1, which seems to encompass all R1a (I have yet to see a single "R1a" that was SRY10831.2- and also M17-. Anyone SRY10831.2- seems to be universally M17+, in other words all R1a are also R1a1). For this reason you will many times see R1a and R1a1 used interchangably within the literature.
One particular group of Y-STR values within R1a shows matches in Central Asia, around the Siberian Altai and Uyghur province of Western China. The recent find of Caucasian mummies in the Takla Makan deserts of the Uyghur province prove that a race of red and blond haired people with Scandinavian features, over 6' tall, once lived in this region. R1a is found at very high percentages in Western Norway, where it reaches frequencies between 23% to 30%. Some researchers believe the Icelandic Sagas, which describe a migration of a population from Asia beyond the Ural mountains, to Norway, may actually be based in fact. Thor Heyerdahl, of Kon Tiki fame, spent the remaining years of his life attempting to prove this theory -- and DNA evidence is seeming to prove him right. The Swedes have long believed this legend, and the emergence of a specific type of Scandinavian R1a with a Central Asian motif seems to support this account.
The MacDonalds have determined that their progenitor, Somerled, belonged to haplogroup R1a (of the same Central Asian motif) and the Douglas's progenitor, Willem de Douglas, recently turned up R1a according to the Clan Douglas DNA project. It seems this holds true for most of the pseudo-aristocracy of Scandinavia. R1a is found at levels of less than 1% in most regions of Ireland, and at levels of 3-5% in England, and only slightly higher in Scotland. The highest concentrations of this haplogroup are seen in areas of Britain colonized by the Norse Vikings. One of the leading DNA experts has called R1a the only sure proof of Norse Viking origins when seen in men of deep British ancestry.
R1b - The most common haplogroup in Western Europe and Britain. This type is believed to have gestated in Spain during the Ice Age, migrating back into Europe with the retreat of the glaciers. People of Celtic ancestry show very close matches with the Basque people of Spain (more recent theories have divided R1b into "Iberian and Non-Iberian" and seem to indicate that some R1b spent the glacial period somewhere outside of Spain). Most people of European or British ancestry will fall into this haplogroup.
R1b is characterized by the P25 mutation (P25+). However, almost all R1b types fall within one of the sub-categories of R1b, typically M269+ called R1b3 (YCC 2003 Chart) or R1b1c (YCC 2005 Chart). This is a rapidly changing group and new SNP's are being discovered regularly so nothing here can be taken as static since it is very much fluid.
It is expected that most of the members of the Grant DNA Project will be of this haplogroup. In fact, the one line we have identified in our project as being the Chiefly line is R1b, which would be expected if the line were related to Andrew Stewart. The Royal line of the Stewarts in Scotland were descended from a family of Breton (in Western France) nobles, so it follows that if Andrew Stewart were of the Royal Line and married into the family, we should expect that the Chiefly line would be R1b. We have also identified a few kits in the project whose owners are directly related to the Chiefly line. [interpretations under review by the Court]
However, most of those that fall into the R1b haplogroup will be unrelated to either the current line of the Chiefs or the original Norse (Haplogroup R1a) line. These DNA signatures represent the various tacksmen, farmers/crofters, peasant classes, etc, that settled in Strathspey or Glenmoriston and assumed the name of the Chief on whose lands they were living. This is the reason that so many project members show very close matches with other individuals, of other surnames, even at the 25 or 37 marker levels in Family Tree DNA's "Y-Search" database.
DNA Bioholograms & Wave Genetics

Spiritual Science: DNA is influneced by words and frequencies
By Grazyna Fosar and Franz Bludorf
DNA Can Be Influenced And Reprogrammed By Words And Frequencies Russian DNA Discoveries
The human DNA is a biological Internet and superior in many aspects to the artificial one. The latest Russian scientific research directly or indirectly explains phenomena such as clairvoyance, intuition, spontaneous and remote acts of healing, self healing, affirmation techniques, unusual light/auras around people (namely spiritual masters), the mindï's influence on weather patterns and much more.
In addition, there is evidence for a whole new type of medicine in which DNA can be influenced and reprogrammed by words and frequencies WITHOUT cutting out and replacing single genes.
Only 10% of our DNA is being used for building proteins. It is this subset of DNA that is of interest to western researchers and is being examined and categorized. The other 90% are considered "junk DNA. The Russian researchers, however, convinced that nature was not dumb, joined linguists and geneticists in a venture to explore those 90% of "junk DNA". Their results, findings and conclusions are simply revolutionary!
According to them, our DNA is not only responsible for the construction of our body but also serves as data storage and communication. The Russian linguists found that the genetic code, especially in the apparently useless 90%, follows the same rules as all our human languages. To this end they compared the rules of syntax (the way in which words are put together to form phrases and sentences), semantics (the study of meaning in language forms) and the basic rules of grammar.
They found that the alkalines of our DNA follow regular grammar and do have set rules just like our languages. So human languages did not appear coincidentally but are a reflection of our inherent DNA.
The Russian biophysicist and molecular biologist Pjotr Garjajev and his colleagues also explored the vibrational behavior of the DNA. [For the sake of brevity I will give only a summary here. For further exploration please refer to the appendix at the end of this article.]
The bottom line was:
"Living chromosomes function just like solitonic/holographic computers using the endogenous DNA laser radiation."
This means that they managed, for example, to modulate certain frequency patterns onto a laser ray and with it influenced the DNA frequency and thus the genetic information itself. Since the basic structure of DNA-alkaline pairs and of language (as explained earlier) are of the same structure, no DNA decoding is necessary. One can simply use words and sentences of the human language!
This, too, was experimentally proven! Living DNA substance (in living tissue, not in vitro) will always react to language-modulated laser rays and even to radio waves, if the proper frequencies are being used. This finally and scientifically explains why affirmations, autogenous training, hypnosis and the like can have such strong effects on humans and their bodies. It is entirely normal and natural for our DNA to react to language. While western researcher cut single genes from the DNA strands and insert them elsewhere, the Russians enthusiastically worked on devices that can influence the cellular metabolism through suitable modulated radio and light frequencies and thus repair genetic defects.
Garjajev's research group succeeded in proving that with this method chromosomes damaged by x-rays for example can be repaired. They even captured information patterns of a particular DNA and transmitted it onto another, thus reprogramming cells to another genome. So they successfully transformed, for example, frog embryos to salamander embryos simply by transmitting the DNA information patterns!
DNA: is also an organic superconductor that can work at normal body temperature. Artificial superconductors require extremely low temperatures of between 200 and 140 C to function.
All superconductors are able to store light and thus information. This is a further explanation of how the DNA can store information. There is another phenomenon linked to DNA and wormholes. Normally, these super small wormholes are highly unstable and are maintained only for the tiniest fractions of a second. Under certain conditions (read about it in the Fosar/Bludorf book above) stable wormholes can organize themselves which then form distinctive vacuum domains in which, for example, gravity can transform into electricity. Vacuum domains are self-radiant balls of ionized gas that contain considerable amounts of energy.
By Grazyna Fosar and Franz Bludorf
DNA Can Be Influenced And Reprogrammed By Words And Frequencies Russian DNA Discoveries
The human DNA is a biological Internet and superior in many aspects to the artificial one. The latest Russian scientific research directly or indirectly explains phenomena such as clairvoyance, intuition, spontaneous and remote acts of healing, self healing, affirmation techniques, unusual light/auras around people (namely spiritual masters), the mindï's influence on weather patterns and much more.
In addition, there is evidence for a whole new type of medicine in which DNA can be influenced and reprogrammed by words and frequencies WITHOUT cutting out and replacing single genes.
Only 10% of our DNA is being used for building proteins. It is this subset of DNA that is of interest to western researchers and is being examined and categorized. The other 90% are considered "junk DNA. The Russian researchers, however, convinced that nature was not dumb, joined linguists and geneticists in a venture to explore those 90% of "junk DNA". Their results, findings and conclusions are simply revolutionary!
According to them, our DNA is not only responsible for the construction of our body but also serves as data storage and communication. The Russian linguists found that the genetic code, especially in the apparently useless 90%, follows the same rules as all our human languages. To this end they compared the rules of syntax (the way in which words are put together to form phrases and sentences), semantics (the study of meaning in language forms) and the basic rules of grammar.
They found that the alkalines of our DNA follow regular grammar and do have set rules just like our languages. So human languages did not appear coincidentally but are a reflection of our inherent DNA.
The Russian biophysicist and molecular biologist Pjotr Garjajev and his colleagues also explored the vibrational behavior of the DNA. [For the sake of brevity I will give only a summary here. For further exploration please refer to the appendix at the end of this article.]
The bottom line was:
"Living chromosomes function just like solitonic/holographic computers using the endogenous DNA laser radiation."
This means that they managed, for example, to modulate certain frequency patterns onto a laser ray and with it influenced the DNA frequency and thus the genetic information itself. Since the basic structure of DNA-alkaline pairs and of language (as explained earlier) are of the same structure, no DNA decoding is necessary. One can simply use words and sentences of the human language!
This, too, was experimentally proven! Living DNA substance (in living tissue, not in vitro) will always react to language-modulated laser rays and even to radio waves, if the proper frequencies are being used. This finally and scientifically explains why affirmations, autogenous training, hypnosis and the like can have such strong effects on humans and their bodies. It is entirely normal and natural for our DNA to react to language. While western researcher cut single genes from the DNA strands and insert them elsewhere, the Russians enthusiastically worked on devices that can influence the cellular metabolism through suitable modulated radio and light frequencies and thus repair genetic defects.
Garjajev's research group succeeded in proving that with this method chromosomes damaged by x-rays for example can be repaired. They even captured information patterns of a particular DNA and transmitted it onto another, thus reprogramming cells to another genome. So they successfully transformed, for example, frog embryos to salamander embryos simply by transmitting the DNA information patterns!
DNA: is also an organic superconductor that can work at normal body temperature. Artificial superconductors require extremely low temperatures of between 200 and 140 C to function.
All superconductors are able to store light and thus information. This is a further explanation of how the DNA can store information. There is another phenomenon linked to DNA and wormholes. Normally, these super small wormholes are highly unstable and are maintained only for the tiniest fractions of a second. Under certain conditions (read about it in the Fosar/Bludorf book above) stable wormholes can organize themselves which then form distinctive vacuum domains in which, for example, gravity can transform into electricity. Vacuum domains are self-radiant balls of ionized gas that contain considerable amounts of energy.
BALL - LIGHTNING
I. Miller, (c) 2000
The soul abides deep in the heart of the flesh,
Where Body and Spirit long to mingle and mesh.
It forms one willing captive amid the silken pillows,
Twists and writhes as undulating waves and billows.
Talent is a monster fed only by Arts,
Embodied as form, its Splendor imparts.
That narcotic need makes our decisions
Glorious, without final cut or revisions.
In a changing world we have music, art and love
To meld or ground dark and light from above.
Hot desire burns omniscient within the senses,
Weaving a spell of impassioned perfect tenses.
That divine response reflected back from our desire
Is the sole motivation for us to continue to aspire.
It is a resigned, passive act merely to believe;
Only the active stroke of Will can ease or relieve.
Finding us bored stiff, I'll see that and raise you One.
A cone of power the likes of which was never done.
Nothing matters to Lovers but sex, death, and religion
All of which reside far beyond gates of human division.
God entreats Goddess, becoming evermore emphatic;
Both now One, growing steadily more empathic.
Power and Light unleashed more awesome than frightning,
A spiralling energy known as Ball-Lightning.
I. Miller, (c) 2000
The soul abides deep in the heart of the flesh,
Where Body and Spirit long to mingle and mesh.
It forms one willing captive amid the silken pillows,
Twists and writhes as undulating waves and billows.
Talent is a monster fed only by Arts,
Embodied as form, its Splendor imparts.
That narcotic need makes our decisions
Glorious, without final cut or revisions.
In a changing world we have music, art and love
To meld or ground dark and light from above.
Hot desire burns omniscient within the senses,
Weaving a spell of impassioned perfect tenses.
That divine response reflected back from our desire
Is the sole motivation for us to continue to aspire.
It is a resigned, passive act merely to believe;
Only the active stroke of Will can ease or relieve.
Finding us bored stiff, I'll see that and raise you One.
A cone of power the likes of which was never done.
Nothing matters to Lovers but sex, death, and religion
All of which reside far beyond gates of human division.
God entreats Goddess, becoming evermore emphatic;
Both now One, growing steadily more empathic.
Power and Light unleashed more awesome than frightning,
A spiralling energy known as Ball-Lightning.